Identification of the prognostic signature based on genomic instability-related alternative splicing in colorectal cancer and its regulatory network
- PMID: 35923577
- PMCID: PMC9340224
- DOI: 10.3389/fbioe.2022.841034
Identification of the prognostic signature based on genomic instability-related alternative splicing in colorectal cancer and its regulatory network
Abstract
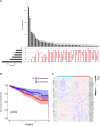
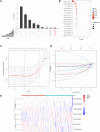
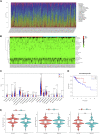
Background: Colorectal cancer (CRC) is a heterogeneous disease with many somatic mutations defining its genomic instability. Alternative Splicing (AS) events, are essential for maintaining genomic instability. However, the role of genomic instability-related AS events in CRC has not been investigated. Methods: From The Cancer Genome Atlas (TCGA) program, we obtained the splicing profiles, the single nucleotide polymorphism, transcriptomics, and clinical information of CRC. Combining somatic mutation and AS events data, a genomic instability-related AS signature was constructed for CRC. Mutations analyses, clinical stratification analyses, and multivariate Cox regression analyses evaluated this signature in training set. Subsequently, we validated the sensitivity and specificity of this prognostic signature using a test set and the entire TCGA dataset. We constructed a nomogram for the prognosis prediction of CRC patients. Differentially infiltrating immune cells were screened by using CIBERSORT. Inmmunophenoscore (IPS) analysis was used to evaluate the response of immunotherapy. The AS events-related splicing factors (SF) were analyzed by Pearson's correlation. The effects of SF regulating the prognostic AS events in proliferation and migration were validated in Caco2 cells. Results: A prognostic signature consisting of seven AS events (PDHA1-88633-ES, KIAA1522-1632-AP, TATDN1-85088-ES, PRMT1-51042-ES, VEZT-23786-ES, AIG1-77972-AT, and PHF11-25891-AP) was constructed. Patients in the high-risk score group showed a higher somatic mutation. The genomic instability risk score was an independent variable associated with overall survival (OS), with a hazard ratio of a risk score of 1.537. The area under the curve of receiver operator characteristic curve of the genomic instability risk score in predicting the OS of CRC patients was 0.733. Furthermore, a nomogram was established and could be used clinically to stratify patients to predict prognosis. Patients defined as high-risk by this signature showed a lower proportion of eosinophils than the low-risk group. Patients with low risk were more sensitive to anti-CTLA4 immunotherapy. Additionally, HSPA1A and FAM50B were two SF regulating the OS-related AS. Downregulation of HSPA1A and FAM50B inhibited the proliferation and migration of Caco2 cells. Conclusion: We constructed an ideal prognostic signature reflecting the genomic instability and OS of CRC patients. HSPA1A and FAM50B were verified as two important SF regulating the OS-related AS.
Keywords: alternative splicing; colorectal cancer; genomic instability; overall survival; splicing factor.
Copyright © 2022 Ding, Hou, Zhao, Chen, Zhao and Xiang.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures






Similar articles
-
Alternative Splicing Events and Their Clinical Significance in Colorectal Cancer: Targeted Therapeutic Opportunities.Cancers (Basel). 2023 Aug 7;15(15):3999. doi: 10.3390/cancers15153999. Cancers (Basel). 2023. PMID: 37568815 Free PMC article. Review.
-
Identification of Mutator-Derived Alternative Splicing Signatures of Genomic Instability for Improving the Clinical Outcome of Cholangiocarcinoma.Front Oncol. 2021 May 14;11:666847. doi: 10.3389/fonc.2021.666847. eCollection 2021. Front Oncol. 2021. PMID: 34055632 Free PMC article.
-
Novel prognostic alternative splicing events in colorectal Cancer: Impact on immune infiltration and therapy response.Int Immunopharmacol. 2024 Sep 30;139:112603. doi: 10.1016/j.intimp.2024.112603. Epub 2024 Jul 22. Int Immunopharmacol. 2024. PMID: 39043103
-
Identification and validation of an immune prognostic signature in colorectal cancer.Int Immunopharmacol. 2020 Nov;88:106868. doi: 10.1016/j.intimp.2020.106868. Epub 2020 Aug 6. Int Immunopharmacol. 2020. PMID: 32771948
-
Cancer Genomic Profiling in Colorectal Cancer: Current Challenges in Subtyping Colorectal Cancers Based on Somatic and Germline Variants.J Anus Rectum Colon. 2021 Jul 29;5(3):213-228. doi: 10.23922/jarc.2021-009. eCollection 2021. J Anus Rectum Colon. 2021. PMID: 34395933 Free PMC article. Review.
Cited by
-
A gene signature related to programmed cell death to predict immunotherapy response and prognosis in colon adenocarcinoma.PeerJ. 2025 Feb 10;13:e18895. doi: 10.7717/peerj.18895. eCollection 2025. PeerJ. 2025. PMID: 39950044 Free PMC article.
-
Alternative Splicing Events and Their Clinical Significance in Colorectal Cancer: Targeted Therapeutic Opportunities.Cancers (Basel). 2023 Aug 7;15(15):3999. doi: 10.3390/cancers15153999. Cancers (Basel). 2023. PMID: 37568815 Free PMC article. Review.
-
Constructing a mitochondrial-related genes model based on machine learning for predicting the prognosis and therapeutic effect in colorectal cancer.Discov Oncol. 2025 May 3;16(1):661. doi: 10.1007/s12672-025-02462-x. Discov Oncol. 2025. PMID: 40317411 Free PMC article.
-
Unveiling prognostic value of JAK/STAT signaling pathway related genes in colorectal cancer: a study of Mendelian randomization analysis.Infect Agent Cancer. 2025 Feb 7;20(1):9. doi: 10.1186/s13027-025-00640-8. Infect Agent Cancer. 2025. PMID: 39920741 Free PMC article.
References
LinkOut - more resources
Full Text Sources
Research Materials

