Prognostic Model and Nomogram Construction and Validation With an Autophagy-Related Gene Signature in Low-Grade Gliomas
- PMID: 35923699
- PMCID: PMC9342864
- DOI: 10.3389/fgene.2022.905751
Prognostic Model and Nomogram Construction and Validation With an Autophagy-Related Gene Signature in Low-Grade Gliomas
Abstract
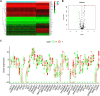
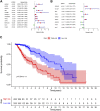
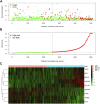
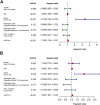
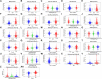
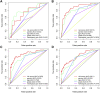
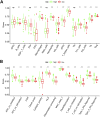
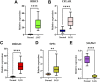
Background : Autophagy plays a vital role in cancer development. However, the prognostic value of autophagy-related genes (ARGs) in low-grade gliomas (LGG) is unclear. This research aimed to investigate whether ARGs correlated with overall survival (OS) in LGG patients. Methods: RNA-sequencing data were obtained from The Cancer Genome Atlas (TCGA) TARGET GTEx database. Gene Ontology and Kyoto Encyclopedia of Genes and Genomes enrichment analysis of ARGs were performed by the "clusterprofile" R package. Cox regression with the wald χ2 test was employed to identify prognostic significant ARGs. Next, the receiver operator characteristic curves were established to evaluate the feasibility of risk score ( ) and other clinical risk factors to predict prognosis. A nomogram was constructed. Correlations between clinical features and ARGs were further verified by a t-test or Kruskal-Wallis test. In addition, the correlations between autophagy and immune cells were assessed through the single-sample gene set enrichment analysis (ssGSEA) and tumor immune estimation resource database. Last, the prediction model was verified by LGG data downloaded from the Chinese Glioma Genome Atlas (CGGA) database. Results: Overall, 35 DE-ARGs were identified. Functional enrichment analysis showed that these genes were mainly related to oxidative stress and regulation of autophagy. Nine ARGs (BAX, BIRC5, CFLAR, DIRAS3, GRID2, MAPK9, MYC, PTK6, and TP53) were significantly associated with OS. Age (Hazard ratio (HR) = 1.063, 95% CI: 1.046-1.080), grade (HR = 3.412, 95% CI: 2.164-5.379), histological type (HR = 0.556, 95% CI: 0.346-0.893), and risk score (HR = 1.135, 95% CI: 1.104-1.167) were independent prognostic risk factors (all p < 0.05). In addition, BIRC5, CFLAR, DIRAS3, TP53, and risk scores were found to correlate significantly with age and tumor grade (all p < 0.05). Immune cell enrichment analysis demonstrated that the types of immune cells and their expression levels in the high-risk group were significantly different from those in the low-risk group (all p < 0.05). A prognostic nomogram was constructed to predict 1-, 3-, and 5-year survival, and the prognostic value of sorted ARGs were verified in the CGGA database and clinical samples. Conclusion: Our findings suggest that the 9 DE-ARGs' risk score model could serve as diagnostic and prognostic biomarkers. The prognostic nomograms could be useful for individualized survival prediction and improved treatment strategies.
Keywords: HADb; TCGA TARGET GTEx; autophagy; bioinformatics analysis; low-grade gliomas (LGG); prognosis.
Copyright © 2022 Li, Huang, Zhu, Yu, Feng, Gu, Jiang, Wang and Huang.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures




References
-
- Amaravadi R. K., Kimmelman A. C., Debnath J. (2019). Targeting Autophagy in Cancer: Recent Advances and Future Directions. Cancer Discov. 9 (9), 1167–1181. 10.1158/2159-8290.cd-19-0292 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

