Genetic and Genomic Characterization of a New Beef Cattle Composite Breed (Purunã) Developed for Production in Pasture-Based Systems
- PMID: 35923708
- PMCID: PMC9341487
- DOI: 10.3389/fgene.2022.858970
Genetic and Genomic Characterization of a New Beef Cattle Composite Breed (Purunã) Developed for Production in Pasture-Based Systems
Abstract
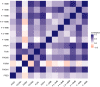
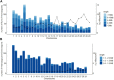
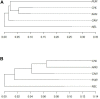
Purunã is a composite beef cattle breed, developed in Southern Brazil by crossing the Angus, Charolais, Canchim, and Caracu breeds. The goal of this study was to perform the first genetic characterization of the Purunã breed, based on both pedigree and genomic information. For this, 100 randomly selected animals were genotyped, and 11,205 animals born from 1997 to 2019 had pedigree information. The genetic analyses performed were principal component analysis, admixture, phylogenic tree, pedigree and genomic inbreeding, linkage disequilibrium (LD), effective population size (Ne), consistency of the gametic phase, runs of homozygosity (ROH), heterozygosity-enriched regions (HERs), and functional analyses of the ROH and HER regions identified. Our findings indicate that Purunã is more genetically related to the Charolais, Canchim, and Angus breeds than Caracu or Nellore. The levels of inbreeding were shown to be small based on all the metrics evaluated and ranged from -0.009 to 0.029. A low (-0.12-0.31) correlation of the pedigree-based inbreeding compared to all the genomic inbreeding coefficients evaluated was observed. The LD average was 0.031 (±0.0517), and the consistency of the gametic phase was shown to be low for all the breed pairs, ranging from 0.42 to 0.27 to the distance of 20 Mb. The Ne values based on pedigree and genomic information were 158 and 115, respectively. A total of 1,839 ROHs were found, and the majority of them are of small length (<4 Mb). An important homozygous region was identified on BTA5 with pathways related to behavioral traits (sensory perception, detection of stimulus, and others), as well as candidate genes related to heat tolerance (MY O 1A), feed conversion rate (RDH5), and reproduction (AMDHD1). A total of 1,799 HERs were identified in the Purunã breed with 92.3% of them classified within the 0.5-1 Mb length group, and 19 HER islands were identified in the autosomal genome. These HER islands harbor genes involved in growth pathways, carcass weight (SDCBP), meat and carcass quality (MT2A), and marbling deposition (CISH). Despite the genetic relationship between Purunã and the founder breeds, a multi-breed genomic evaluation is likely not feasible due to their population structure and low consistency of the gametic phase among them.
Keywords: beef cattle; genomic diversity; inbreeding coefficient; persistency of the gametic phase; runs of heterozygosity; runs of homozygosity.
Copyright © 2022 Mulim, Brito, Batista Pinto, Moletta, Da Silva and Pedrosa.
Conflict of interest statement
Author LD is employed by the company NEOGEN Corporation. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


References
-
- Alemu S. W., Kadri N. K., Harland C., Faux P., Charlier C., Caballero A., et al. (2021). An Evaluation of Inbreeding Measures Using a Whole-Genome Sequenced Cattle Pedigree. Heredity 126, 410–423. 10.1038/s41437-020-00383-9 PubMed Abstract | 10.1038/s41437-020-00383-9 | Google Scholar - DOI - DOI - PMC - PubMed
-
- Alexander D. H., Lange K. (2011). Enhancements to the ADMIXTURE Algorithm for Individual Ancestry Estimation. BMC Bioinforma. 12, 1–6. 10.1186/1471-2105-12-246 PubMed Abstract | 10.1186/1471-2105-12-246 | Google Scholar - DOI - DOI - PMC - PubMed
-
- Alexander D. H., Shringarpure S. S., Novembre J., Lange K. (2015). Admixture 1.3 Software Manual. Cold Spring Harbor Lab. Google Scholar
-
- AmiGO (2021a). AmiGO 2: Term Details for “Detection of Stimulus” (GO:0051606). Available at: http://amigo.geneontology.org/amigo/term/GO:0051606 (Accessed November 18, 2021). Google Scholar
-
- AmiGO (2021b). AmiGO 2: Term Details for “Growth” (GO:0040007). Available at: http://amigo.geneontology.org/amigo/term/GO:0040007 (Accessed November 24, 2021). Google Scholar
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

