GWAS and RNA-seq analysis uncover candidate genes associated with alkaline stress tolerance in maize (Zea mays L.) seedlings
- PMID: 35923879
- PMCID: PMC9340071
- DOI: 10.3389/fpls.2022.963874
GWAS and RNA-seq analysis uncover candidate genes associated with alkaline stress tolerance in maize (Zea mays L.) seedlings
Abstract
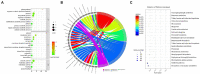
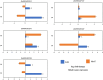
Soil salt-alkalization is a common yet critical environmental stress factor for plant growth and development. Discovering and exploiting genes associated with alkaline tolerance in maize (Zea mays L.) is helpful for improving alkaline resistance. Here, an association panel consisting of 200 maize lines was used to identify the genetic loci responsible for alkaline tolerance-related traits in maize seedlings. A total of nine single-nucleotide polymorphisms (SNPs) and their associated candidate genes were found to be significantly associated with alkaline tolerance using a genome-wide association study (GWAS). An additional 200 genes were identified when the screen was extended to include a linkage disequilibrium (LD) decay distance of r2 ≥ 0.2 from the SNPs. RNA-sequencing (RNA-seq) analysis was then conducted to confirm the linkage between the candidate genes and alkali tolerance. From these data, a total of five differentially expressed genes (DEGs; |log2FC| ≥ 0.585, p < 0.05) were verified as the hub genes involved in alkaline tolerance. Subsequently, two candidate genes, Zm00001d038250 and Zm00001d001960, were verified to affect the alkaline tolerance of maize seedlings by qRT-PCR analysis. These genes were putatively involved protein binding and "flavonoid biosynthesis process," respectively, based on Kyoto Encyclopedia of Genes and Genomes (KEGG) and Gene Ontology (GO) enrichment analyses. Gene promoter region contains elements related to stress and metabolism. The results of this study will help further elucidate the mechanisms of alkaline tolerance in maize, which will provide the groundwork for future breeding projects.
Keywords: RNA-seq; alkali tolerance; candidate genes; genome-wide association study; maize.
Copyright © 2022 Li, Jia, Zhou, Liu, Cao, Zhou, Wang and Di.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures





References
-
- Ambede J. G., Netondo G. W., Mwai G. N., Musyimi M. (2012). NaCl salinity affects germination, growth, physiology, and biochemistry of bambara groundnut. Brazilian J. Plant Physiol. 24, 151–160. doi: 10.1590/S1677-04202012000300002 - DOI
-
- Bekh-Ochir D., Shimada S., Yamagami A., Kanda S., Ogawa K., Nakazawa M., et al. (2013). A novel mitochondrial DnaJ/Hsp40 family protein BIL2 promotes plant growth and resistance against environmental stress in brassinosteroid signaling. Planta 237, 1509–1525. doi: 10.1007/s00425-013-1859-3, PMID: - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Research Materials

