A Novel Prognostic Risk Model for Cervical Cancer Based on Immune Checkpoint HLA-G-Driven Differentially Expressed Genes
- PMID: 35924232
- PMCID: PMC9341272
- DOI: 10.3389/fimmu.2022.851622
A Novel Prognostic Risk Model for Cervical Cancer Based on Immune Checkpoint HLA-G-Driven Differentially Expressed Genes
Abstract
Human leukocyte antigen G (HLA-G) is a potential checkpoint molecule that plays a key role in cervical carcinogenesis. The purpose of this study was to construct and validate a prognostic risk model to predict the overall survival (OS) of cervical cancer patients, providing a reference for individualized clinical treatment that may lead to better clinical outcomes. HLA-G-driven differentially expressed genes (DEGs) were obtained from two cervical carcinoma cell lines, namely, SiHa and HeLa, with stable overexpression of HLA-G by RNA sequencing (RNA-seq). The biological functions of these HLA-G-driven DEGs were analysed by GO enrichment and KEGG pathway using the "clusterProfiler" package. The protein-protein interactions (PPIs) were assessed using the STRING database. The prognostic relevance of each DEG was evaluated by univariate Cox regression using the TCGA-CESC dataset. After the TCGA-CESC cohort was randomly divided into training set and testing set, and a prognostic risk model was constructed by LASSO and stepwise multivariate Cox regression analysis in training set and validated in testing set or in different types of cervical cancer set. The predictive ability of the prognostic risk model or nomogram was evaluated by a series of bioinformatics methods. A total of 1108 candidate HLA-G-driven DEGs, including 391 upregulated and 717 downregulated genes, were obtained and were enriched mostly in the ErbB pathway, steroid biosynthesis, and MAPK pathway. Then, an HLA-G-driven DEG signature consisting of the eight most important prognostic genes CD46, LGALS9, PGM1, SPRY4, CACNB3, PLIN2, MSMO1, and DAGLB was identified as a key predictor of cervical cancer. Multivariate Cox regression analysis showed that this signature is an independent risk factor for the overall survival of CESC patients. Kaplan-Meier survival analysis showed that the 5-year overall survival rate is 23.0% and 84.6% for the high-risk and low-risk patients, respectively (P<0.001). The receiver operating characteristic (ROC) curve of this prognostic model with an area under the curve (AUC) was 0.896 for 5 years, which was better than that of other clinical traits. This prognostic risk model was also successfully validated in different subtypes of cervical cancer, including the keratinizing squamous cell carcinoma, non-keratinizing squamous cell carcinoma, squamous cell neoplasms, non-squamous cell neoplasms set. Single-sample gene set enrichment (ssGSEA) algorithm and Tumor Immune Dysfunction and Exclusion (TIDE) analysis confirmed that this signature influence tumour microenvironment and immune checkpoint blockade. A nomogram that integrated risk score, age, clinical stage, histological grade, and pathological type was then built to predict the overall survival of CESC patients and evaluated by calibration curves, AUC, concordance index (C-index) and decision curve analysis (DCA). To summarize, we developed and validated a novel prognostic risk model for cervical cancer based on HLA-G-driven DEGs, and the prognostic signature showed great ability in predicting the overall survival of patients with cervical cancer.
Keywords: HLA-G; cervical cancer; immune checkpoint; mRNA signature; prediction; prognosis.
Copyright © 2022 Xu, Wang, Xing and Wang.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
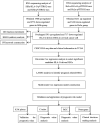
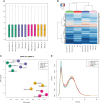
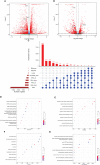
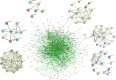


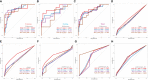


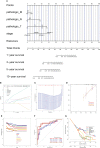

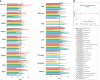
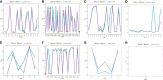

References
-
- WHO . Cervical Cancer . World Health Organization. Available at: https://www.who.int/news-room/fact-sheets/detail/human-papillomavirus-(h... (Accessed September 21, 2020).
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

