Integrated analysis of the functions and clinical implications of exosome circRNAs in colorectal cancer
- PMID: 35924235
- PMCID: PMC9339618
- DOI: 10.3389/fimmu.2022.919014
Integrated analysis of the functions and clinical implications of exosome circRNAs in colorectal cancer
Abstract
Background: Exosome circRNAs (Exo-circRNAs) regulate cancer progression and intercellular crosstalk in the tumor microenvironment. However, their biological functions and potential clinical importance in colorectal cancer (CRC) remain unknown.
Methods: We used exoRBase 2.0 data to identify significant differentially expressed Exo-circRNAs (Exo-DEcircRNAs) in CRC patients and healthy individuals. The least absolute shrinkage and selector operation algorithm, support vector machine-recursive feature elimination, and multivariate Cox regression analyses were used to select candidate Exo-circRNAs and constructed a diagnostic model. Quantitative reverse transcription-polymerase chain reaction analysis was performed to confirm the expression of Exo-circRNAs in the serum samples of patients. Furthermore, we constructed an exosome circRNA-miRNA-mRNA network for CRC. Upregulated target mRNAs in the exosome competing endogenous RNA (Exo-ceRNA) network were used for functional and pathway enrichment analyses. We identified 22 immune cell types in CRC patients using CIBERSORT. Correlation analysis revealed the relationship between Exo-ceRNA networks and immune-infiltrating cells. The relationship between target mRNAs and immunotherapeutic response was also explored. Finally, using the Kaplan-Meier survival curve, a prognostic upregulated target mRNA was screened. We constructed a survival-related Exo-ceRNA subnetwork and explored the correlation between the Exo-ceRNA subnetwork and immune-infiltrating cells.
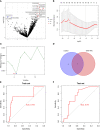
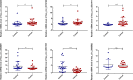
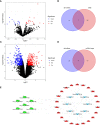
Results: The constructed diagnostic model had a high area under the curve (AUC) value in both the training and validation sets (AUC = 0.744 and AUC = 0.741, respectively). qRT-PCR confirmed that the Exo-circRNAs were differentially expressed in CRC serum samples. We constructed Exo-ceRNA networks based on the interactions among seven upregulated Exo-DEcircRNAs, eight differentially expressed miRNAs, and twenty-two differentially expressed mRNAs in CRC. Functional enrichment analysis revealed that the upregulated target mRNAs were significantly enriched in cytoskeletal motor activity and the PI3K-Akt signaling pathway. Co-expression analysis showed a significant correlation between the Exo-ceRNA networks and immune cells. The significant correlation was observed between target mRNAs and the immunotherapeutic response. Additionally, based on the prognostic upregulated target gene (RGS2), we constructed a survival-related Exo-ceRNA subnetwork (Exosome hsa_circ_0050334-hsa_miR_182_5p-RGS2). CIBERSORT results revealed that the Exo-ceRNA subnetwork correlated with M2 macrophages (P = 4.6e-07, R = 0.31).
Conclusions: Our study identified an Exo-diagnostic model, established Exo-ceRNA networks, and explored the correlation between Exo-ceRNA networks and immune cell infiltration in CRC. These findings elucidated the biological functions of Exo-circRNAs and their potential clinical implications.
Keywords: circRNA; colorectal cancer; competing endogenous RNA network; diagnostic model; exosome; immune-infiltrating cell.
Copyright © 2022 Lei, Zhang, Wang, Liu, Feng and Song.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



Similar articles
-
Identification of Serum Exosome-Derived circRNA-miRNA-TF-mRNA Regulatory Network in Postmenopausal Osteoporosis Using Bioinformatics Analysis and Validation in Peripheral Blood-Derived Mononuclear Cells.Front Endocrinol (Lausanne). 2022 Jun 9;13:899503. doi: 10.3389/fendo.2022.899503. eCollection 2022. Front Endocrinol (Lausanne). 2022. PMID: 35757392 Free PMC article.
-
Whole-transcriptome analysis reveals a potential hsa_circ_0001955/hsa_circ_0000977-mediated miRNA-mRNA regulatory sub-network in colorectal cancer.Aging (Albany NY). 2020 Mar 28;12(6):5259-5279. doi: 10.18632/aging.102945. Epub 2020 Mar 28. Aging (Albany NY). 2020. PMID: 32221048 Free PMC article.
-
Construction of endogenous RNA regulatory network for colorectal cancer based on bioinformatics.Zhong Nan Da Xue Xue Bao Yi Xue Ban. 2022 Apr 28;47(4):416-430. doi: 10.11817/j.issn.1672-7347.2022.210532. Zhong Nan Da Xue Xue Bao Yi Xue Ban. 2022. PMID: 35545337 Free PMC article. Chinese, English.
-
The circRNA-miRNA-mRNA regulatory network in plasma and peripheral blood mononuclear cells and the potential associations with the pathogenesis of systemic lupus erythematosus.Clin Rheumatol. 2023 Jul;42(7):1885-1896. doi: 10.1007/s10067-023-06560-5. Epub 2023 Mar 2. Clin Rheumatol. 2023. PMID: 36862342 Review.
-
Biological functions and molecular mechanisms of exosome-derived circular RNAs and their clinical implications in digestive malignancies: the vintage in the bottle.Ann Med. 2024 Dec;56(1):2420861. doi: 10.1080/07853890.2024.2420861. Epub 2024 Nov 1. Ann Med. 2024. PMID: 39484707 Free PMC article. Review.
Cited by
-
Progress of regulatory RNA in small extracellular vesicles in colorectal cancer.Front Cell Dev Biol. 2023 Jul 12;11:1225965. doi: 10.3389/fcell.2023.1225965. eCollection 2023. Front Cell Dev Biol. 2023. PMID: 37519298 Free PMC article. Review.
-
Gene Coexpression and miRNA Regulation: A Path to Early Intervention in Colorectal Cancer.Hum Gene Ther. 2024 Oct;35(19-20):855-867. doi: 10.1089/hum.2023.207. Epub 2024 Jul 4. Hum Gene Ther. 2024. PMID: 38767504
-
Exosomal circRNAs: Deciphering the novel drug resistance roles in cancer therapy.J Pharm Anal. 2025 Feb;15(2):101067. doi: 10.1016/j.jpha.2024.101067. Epub 2024 Aug 13. J Pharm Anal. 2025. PMID: 39957900 Free PMC article. Review.
-
Mechanism of LINC01018/miR-182-5p/Rab27B in the immune escape through PD-L1-mediated CD8+ T cell suppression in glioma.Biol Direct. 2025 May 21;20(1):61. doi: 10.1186/s13062-025-00651-w. Biol Direct. 2025. PMID: 40399992 Free PMC article.
-
Extracellular Vesicle Preparation and Analysis: A State-of-the-Art Review.Adv Sci (Weinh). 2024 Aug;11(30):e2401069. doi: 10.1002/advs.202401069. Epub 2024 Jun 14. Adv Sci (Weinh). 2024. PMID: 38874129 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

