Oral Supplementation of Houttuynia cordata Extract Reduces Viremia in PRRSV-1 Modified-Live Virus-Vaccinated Pigs in Response to the HP-PRRSV-2 Challenge
- PMID: 35924249
- PMCID: PMC9339630
- DOI: 10.3389/fimmu.2022.929338
Oral Supplementation of Houttuynia cordata Extract Reduces Viremia in PRRSV-1 Modified-Live Virus-Vaccinated Pigs in Response to the HP-PRRSV-2 Challenge
Abstract
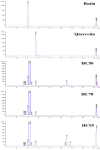
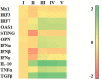
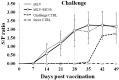
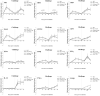
This study evaluated the in vitro antiviral activities and the ex vivo immunomodulatory effects of Houttuynia cordata Thunb. (HC) ethanolic extracts in response to porcine reproductive and respiratory syndrome virus (PRRSV). In addition, this study evaluated the in vivo effects of oral supplementation of HC extract on immune responses to and cross-protective efficacy of PRRSV-1 modified-live virus (MLV) vaccine against the highly pathogenic (HP)-PRRSV-2 challenge. In vitro experiments demonstrated that HC extracted in either 50%, 70%, or 95% ethanol (referred to as HC50, HC70, and HC95, respectively) significantly interfered with PRRSV replication in MARC-145 cells. Ex vivo experiments revealed that all HC extracts significantly enhanced mRNA expressions of type I interferon-regulated genes, type I and II interferon (IFN), and pro- and anti-inflammatory cytokines in HP-PRRSV-2-inoculated monocyte-derived macrophages. An in vivo experiment included four groups of six pigs (4 weeks old; n = 24). Group 1 and group 2 were vaccinated with the PRRSV-1 MLV vaccine at 0 dpv (day post vaccination). Group 2 also received oral administration of HC50 extract at 0-49 dpv. Group 3 received the PRRSV-1 MLV vaccine solvent at 0 dpv, while group 4 served as strict control. Groups 1-3 were challenged intranasally with HP-PRRSV-2 at 28 dpv and immune-related and clinical parameters were monitored weekly until 49 dpv. Compared to group 1, group 2 demonstrated significantly increased IFN regulatory factor 3 mRNA expression of PRRSV-recalled peripheral blood mononuclear cells, and significantly reduced HP-PRRSV-2 viremia. No difference in PRRSV-specific antibody responses, rectal temperature, clinical scores, and average daily weight gain was detected. Our study reports the immunomodulatory and anti-PRRSV potentials of HC extract in PRRSV-1 MLV-vaccinated/HP-PRRSV-2 challenged pigs.
Keywords: cross-protection; houttuynia cordata; interferon; modified-live virus vaccine; porcine reproductive and respiratory syndrome virus.
Copyright © 2022 Ruansit and Charerntantanakul.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

Similar articles
-
Oral supplementation of quercetin in PRRSV-1 modified-live virus vaccinated pigs in response to HP-PRRSV-2 challenge.Vaccine. 2020 Apr 23;38(19):3570-3581. doi: 10.1016/j.vaccine.2020.03.019. Epub 2020 Mar 14. Vaccine. 2020. PMID: 32184034
-
Co-administration of saponin quil A and PRRSV-1 modified-live virus vaccine up-regulates gene expression of type I interferon-regulated gene, type I and II interferon, and inflammatory cytokines and reduces viremia in response to PRRSV-2 challenge.Vet Immunol Immunopathol. 2018 Nov;205:24-34. doi: 10.1016/j.vetimm.2018.10.005. Epub 2018 Oct 24. Vet Immunol Immunopathol. 2018. PMID: 30458999
-
Immune response and protective efficacy of intramuscular and intradermal vaccination with porcine reproductive and respiratory syndrome virus 1 (PRRSV-1) modified live vaccine against highly pathogenic PRRSV-2 (HP-PRRSV-2) challenge, either alone or in combination with of PRRSV-1.Vet Microbiol. 2020 May;244:108655. doi: 10.1016/j.vetmic.2020.108655. Epub 2020 Mar 27. Vet Microbiol. 2020. PMID: 32402335
-
Pharmacological Effects of Houttuynia cordata Thunb (H. cordata): A Comprehensive Review.Pharmaceuticals (Basel). 2022 Aug 29;15(9):1079. doi: 10.3390/ph15091079. Pharmaceuticals (Basel). 2022. PMID: 36145299 Free PMC article. Review.
-
The therapeutic potential of Houttuynia cordata: A current review.Heliyon. 2022 Aug 24;8(8):e10386. doi: 10.1016/j.heliyon.2022.e10386. eCollection 2022 Aug. Heliyon. 2022. PMID: 36061012 Free PMC article. Review.
Cited by
-
Porcine Reproductive and Respiratory Syndrome Virus: Challenges and Advances in Vaccine Development.Vaccines (Basel). 2025 Feb 28;13(3):260. doi: 10.3390/vaccines13030260. Vaccines (Basel). 2025. PMID: 40266104 Free PMC article. Review.
-
Quality and production enhancement of fish mint, Houttuynia cordata Thunb., cultivated in a hydroponic planting system with designed plant growth-promoting additives.Heliyon. 2024 Mar 26;10(7):e28755. doi: 10.1016/j.heliyon.2024.e28755. eCollection 2024 Apr 15. Heliyon. 2024. PMID: 38586372 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

