Bioinformatic Analysis and Translational Validation of Psoriasis Candidate Genes for Precision Medicine
- PMID: 35924255
- PMCID: PMC9343179
- DOI: 10.2147/CCID.S378143
Bioinformatic Analysis and Translational Validation of Psoriasis Candidate Genes for Precision Medicine
Abstract
Background: Psoriasis is a recurrent, chronic, inflammation- and immune-mediated skin disease with multiple causative factors. However, the genetic markers associated with recurrence have not yet been fully elucidated. Accordingly, in this study, we aimed to identify markers associated with the recurrence of psoriasis.
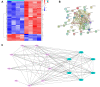
Methods: We analyzed differentially expressed genes to determine which targets were associated with the recurrence of psoriasis and used these data to construct a protein-protein interaction network using Cytoscape software. The results were then validated by analysis of core targets using Gene Expression Omnibus (GEO) datasets and clinical samples. Functional enrichment analysis was used to explore the potential mechanisms mediating the recurrence of psoriasis.
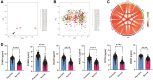
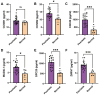
Results: We screened out six core targets that played important roles in recurrence of psoriasis, and validation of GEO datasets and clinical samples showed that the expression levels of five core targets were higher in patients with psoriasis than in healthy individuals. Functional enrichment analysis revealed that the cell cycle and oocyte meiosis signaling pathways were involved in the recurrence of psoriasis.
Conclusion: Our findings provided insights into the mechanisms mediating the onset and recurrence of psoriasis.
Keywords: cell cycle; gene expression profiling; pathogenic mechanism; psoriasis; recurrence.
© 2022 Li et al.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

Similar articles
-
Decoding Psoriasis: Integrated Bioinformatics Approach to Understand Hub Genes and Involved Pathways.Curr Pharm Des. 2020;26(29):3619-3630. doi: 10.2174/1381612826666200311130133. Curr Pharm Des. 2020. PMID: 32160841
-
Integrated bioinformatic analysis of differentially expressed genes and signaling pathways in plaque psoriasis.Mol Med Rep. 2019 Jul;20(1):225-235. doi: 10.3892/mmr.2019.10241. Epub 2019 May 15. Mol Med Rep. 2019. PMID: 31115544 Free PMC article.
-
ZNF384: A Potential Therapeutic Target for Psoriasis and Alzheimer's Disease Through Inflammation and Metabolism.Front Immunol. 2022 May 20;13:892368. doi: 10.3389/fimmu.2022.892368. eCollection 2022. Front Immunol. 2022. PMID: 35669784 Free PMC article.
-
Comparisons of gene expression in normal, lesional, and non-lesional psoriatic skin using DNA microarray techniques.Int J Dermatol. 2014 Oct;53(10):1213-20. doi: 10.1111/ijd.12476. Epub 2014 Jul 11. Int J Dermatol. 2014. PMID: 25041445
-
Network analysis of potential risk genes for psoriasis.Hereditas. 2021 Jun 16;158(1):21. doi: 10.1186/s41065-021-00186-w. Hereditas. 2021. PMID: 34134787 Free PMC article.
Cited by
-
miR-16-5p, miR-21-5p, and miR-155-5p in circulating vesicles as psoriasis biomarkers.Sci Rep. 2025 Feb 26;15(1):6971. doi: 10.1038/s41598-025-91532-9. Sci Rep. 2025. PMID: 40011539 Free PMC article.
References
-
- Boehncke WH, Schön MP. Psoriasis. Lancet. 2015;386(9997):983–994. - PubMed
-
- Lou F, Sun Y, Xu Z, et al. Excessive Polyamine Generation in Keratinocytes Promotes Self-RNA Sensing by Dendritic Cells in Psoriasis. Immunity. 2020;53(1):204–216.e10. - PubMed
-
- Michalek IM, Loring B, John SM. A systematic review of worldwide epidemiology of psoriasis. J Eur Acad Dermatol Venereol. 2017;31(2):205–212. - PubMed
LinkOut - more resources
Full Text Sources

