SAA fibrils involved in AA amyloidosis are similar in bulk and by single particle reconstitution: A MAS solid-state NMR study
- PMID: 35924280
- PMCID: PMC9340516
- DOI: 10.1016/j.yjsbx.2022.100069
SAA fibrils involved in AA amyloidosis are similar in bulk and by single particle reconstitution: A MAS solid-state NMR study
Abstract
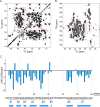
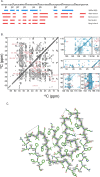
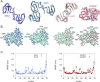
AA amyloidosis is one of the most prevalent forms of systemic amyloidosis and affects both humans and other vertebrates. In this study, we compare MAS solid-state NMR data with a recent cryo-EM study of fibrils involving full-length murine SAA1.1. We address the question whether the specific requirements for the reconstitution of an amyloid fibril structure by cryo-EM can potentially yield a bias towards a particular fibril polymorph. We employ fibril seeds extracted from in to vivo material to imprint the fibril structure onto the biochemically produced protein. Sequential assignments yield the secondary structure elements in the fibril state. Long-range DARR and PAR experiments confirm largely the topology observed in the ex-vivo cryo-EM study. We find that the β-sheets identified in the NMR experiments are similar to the β-sheets found in the cryo-EM study, with the exception of amino acids 33-42. These residues cannot be assigned by solid-state NMR, while they adopt a stable β-sheet in the cryo-EM structure. We suggest that the differences between MAS solid-state NMR and cryo-EM data are a consequence of a second conformer involving residues 33-42. Moreover, we were able to characterize the dynamic C-terminal tail of SAA in the fibril state. The C-terminus is flexible, remains detached from the fibrils, and does not affect the SAA fibril structure as confirmed further by molecular dynamics simulations. As the C-terminus can potentially interact with other cellular components, binding to cellular targets can affect its accessibility for protease digestion.
Keywords: Amyloid fibrils; Cryo electron microscopy (EM); Magic Angle Spinning (MAS); Solid-state NMR; Structure.
© 2022 The Authors.
Conflict of interest statement
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures





Similar articles
-
Cryo-EM and Solid State NMR Together Provide a More Comprehensive Structural Investigation of Protein Fibrils.bioRxiv [Preprint]. 2024 Jun 2:2024.05.30.596698. doi: 10.1101/2024.05.30.596698. bioRxiv. 2024. Update in: Protein Sci. 2024 Oct;33(10):e5168. doi: 10.1002/pro.5168. PMID: 38853912 Free PMC article. Updated. Preprint.
-
Seeded fibrils of the germline variant of human λ-III immunoglobulin light chain FOR005 have a similar core as patient fibrils with reduced stability.J Biol Chem. 2020 Dec 25;295(52):18474-18484. doi: 10.1074/jbc.RA120.016006. Epub 2020 Oct 22. J Biol Chem. 2020. PMID: 33093170 Free PMC article.
-
Cryo-EM demonstrates the in vitro proliferation of an ex vivo amyloid fibril morphology by seeding.Nat Commun. 2022 Jan 10;13(1):85. doi: 10.1038/s41467-021-27688-5. Nat Commun. 2022. PMID: 35013242 Free PMC article.
-
Protein denaturation and aggregation: Cellular responses to denatured and aggregated proteins.Ann N Y Acad Sci. 2005 Dec;1066:181-221. doi: 10.1196/annals.1363.030. Ann N Y Acad Sci. 2005. PMID: 16533927 Review.
-
Methods to study the structure of misfolded protein states in systemic amyloidosis.Biochem Soc Trans. 2021 Apr 30;49(2):977-985. doi: 10.1042/BST20201022. Biochem Soc Trans. 2021. PMID: 33929491 Review.
Cited by
-
Cryo-EM and Solid State NMR Together Provide a More Comprehensive Structural Investigation of Protein Fibrils.bioRxiv [Preprint]. 2024 Jun 2:2024.05.30.596698. doi: 10.1101/2024.05.30.596698. bioRxiv. 2024. Update in: Protein Sci. 2024 Oct;33(10):e5168. doi: 10.1002/pro.5168. PMID: 38853912 Free PMC article. Updated. Preprint.
-
Cryo-EM and solid state NMR together provide a more comprehensive structural investigation of protein fibrils.Protein Sci. 2024 Oct;33(10):e5168. doi: 10.1002/pro.5168. Protein Sci. 2024. PMID: 39276003
References
-
- Andronesi O.C., Becker S., Seidel K., Heise H., Young H.S., Baldus M. Determination of membrane protein structure and dynamics by magic-angle-spinning solid-state NMR spectroscopy. J. Am. Chem. Soc. 2005;127:12965–12974. - PubMed
-
- Baldus MARC, Petkova A.T., Herzfeld JUDITH, Griffin R.G. Cross Polarization in the Tilted Frame: Assignment and Spectral Simplification in Heteronuclear Spin Systems. Mol. Phys. 1998;95(6):1197–1207.
-
- Brunger A.F., Nienhuis H.L.A., Bijzet J., Hazenberg B.P.C. Causes of AA amyloidosis: a systematic review. Amyloid. 2020;27(1):1–12. - PubMed
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

