Investigating Peptidoglycan Recycling Pathways in Tannerella forsythia with N-Acetylmuramic Acid Bioorthogonal Probes
- PMID: 35924866
- PMCID: PMC9464701
- DOI: 10.1021/acsinfecdis.2c00333
Investigating Peptidoglycan Recycling Pathways in Tannerella forsythia with N-Acetylmuramic Acid Bioorthogonal Probes
Abstract
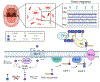
The human oral microbiome is the second largest microbial community in humans, harboring over 700 bacterial species, which aid in digestion and protect from growth of disease-causing pathogens. One such oral pathogen, Tannerella forsythia, along with other species, contributes to the pathogenesis of periodontitis. T. forsythia is unable to produce its own N-acetylmuramic acid (NAM) sugar, essential for peptidoglycan biosynthesis and therefore must scavenge NAM from other species with which it cohabitates. Here, we explore the recycling potential of T. forsythia for NAM uptake with a bioorthogonal modification into its peptidoglycan, allowing for click-chemistry-based visualization of the cell wall structure. Additionally, we identified NAM recycling enzyme homologues in T. forsythia that are similar to the enzymes found in Pseudomonas putida. These homologues were then genetically transformed into a laboratory safe Escherichia coli strain, resulting in the efficient incorporation of unnatural NAM analogues into the peptidoglycan backbone and its visualization, alone or in the presence of human macrophages. This strain will be useful in further studies to probe NAM recycling and peptidoglycan scavenging pathways of T. forsythia and other cohabiting bacteria.
Keywords: N-acetylmuramic acid probes; bacterial cell-wall remodeling; bacterial pathogenesis; bioorthogonal; oral microbiome; peptidoglycan.
Figures

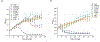
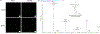


Similar articles
-
Bioorthogonal Labeling and Click-Chemistry-Based Visualization of the Tannerella forsythia Cell Wall.Methods Mol Biol. 2024;2727:1-16. doi: 10.1007/978-1-0716-3491-2_1. Methods Mol Biol. 2024. PMID: 37815704 Free PMC article.
-
Peptidoglycan-type analysis of the N-acetylmuramic acid auxotrophic oral pathogen Tannerella forsythia and reclassification of the peptidoglycan-type of Porphyromonas gingivalis.BMC Microbiol. 2019 Sep 2;19(1):200. doi: 10.1186/s12866-019-1575-7. BMC Microbiol. 2019. PMID: 31477019 Free PMC article.
-
Peptidoglycan synthesis in Tannerella forsythia: Scavenging is the modus operandi.Mol Oral Microbiol. 2018 Apr;33(2):125-132. doi: 10.1111/omi.12210. Epub 2018 Feb 12. Mol Oral Microbiol. 2018. PMID: 29247483 Free PMC article. Review.
-
Utilization of different MurNAc sources by the oral pathogen Tannerella forsythia and role of the inner membrane transporter AmpG.BMC Microbiol. 2020 Nov 17;20(1):352. doi: 10.1186/s12866-020-02006-z. BMC Microbiol. 2020. PMID: 33203363 Free PMC article.
-
Peptidoglycan Salvage Enables the Periodontal Pathogen Tannerella forsythia to Survive within the Oral Microbial Community.Microb Physiol. 2021;31(2):123-134. doi: 10.1159/000516751. Epub 2021 Jun 9. Microb Physiol. 2021. PMID: 34107471 Review.
Cited by
-
Bioorthogonal Labeling and Click-Chemistry-Based Visualization of the Tannerella forsythia Cell Wall.Methods Mol Biol. 2024;2727:1-16. doi: 10.1007/978-1-0716-3491-2_1. Methods Mol Biol. 2024. PMID: 37815704 Free PMC article.
-
Chemical tools to study and modulate glycan-mediated host-bacteria interactions.Curr Opin Chem Biol. 2025 Aug;87:102603. doi: 10.1016/j.cbpa.2025.102603. Epub 2025 Jun 4. Curr Opin Chem Biol. 2025. PMID: 40472591 Review.
-
Minimalist Tetrazine N-Acetyl Muramic Acid Probes for Rapid and Efficient Labeling of Commensal and Pathogenic Peptidoglycans in Living Bacterial Culture and During Macrophage Invasion.J Am Chem Soc. 2024 Mar 13;146(10):6817-6829. doi: 10.1021/jacs.3c13644. Epub 2024 Mar 1. J Am Chem Soc. 2024. PMID: 38427023 Free PMC article.
-
The resuscitation-promoting factor (Rpf) from Micrococcus luteus and its putative reaction product 1,6-anhydro-MurNAc increase culturability of environmental bacteria.Access Microbiol. 2023 Sep 22;5(9):000647.v4. doi: 10.1099/acmi.0.000647.v4. eCollection 2023. Access Microbiol. 2023. PMID: 37841103 Free PMC article.
-
The Legionella pneumophila peptidoglycan recycling kinase, AmgK, is essential for survival and replication inside host alveolar macrophages.bioRxiv [Preprint]. 2025 Mar 22:2025.03.21.644609. doi: 10.1101/2025.03.21.644609. bioRxiv. 2025. PMID: 40166355 Free PMC article. Preprint.
References
-
- Kilian M; Chapple ILC; Hannig M; Marsh PD; Meuric V; Pedersen AML; Tonetti MS; Wade WG; Zaura E, The oral microbiome - an update for oral healthcare professionals. Brit Dent J 2016, 221 (10), 657–666. - PubMed
-
- Tanner ACR; Izard J, Tannerella forsythia, a periodontal pathogen entering the genomic era. Periodontol 2000 2006, 42, 88–113. - PubMed
-
- Tonetti MS; Jepsen S; Jin L; Otomo-Corgel J, Impact of the global burden of periodontal diseases on health, nutrition and wellbeing of mankind: A call for global action. J Clin Periodontol 2017, 44 (5), 456–462. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

