The evolution of a counter-defense mechanism in a virus constrains its host range
- PMID: 35924892
- PMCID: PMC9391042
- DOI: 10.7554/eLife.79549
The evolution of a counter-defense mechanism in a virus constrains its host range
Abstract
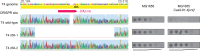
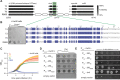
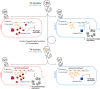
Bacteria use diverse immunity mechanisms to defend themselves against their viral predators, bacteriophages. In turn, phages can acquire counter-defense systems, but it remains unclear how such mechanisms arise and what factors constrain viral evolution. Here, we experimentally evolved T4 phage to overcome a phage-defensive toxin-antitoxin system, toxIN, in Escherichia coli. Through recombination, T4 rapidly acquires segmental amplifications of a previously uncharacterized gene, now named tifA, encoding an inhibitor of the toxin, ToxN. These amplifications subsequently drive large deletions elsewhere in T4's genome to maintain a genome size compatible with capsid packaging. The deleted regions include accessory genes that help T4 overcome defense systems in alternative hosts. Thus, our results reveal a trade-off in viral evolution; the emergence of one counter-defense mechanism can lead to loss of other such mechanisms, thereby constraining host range. We propose that the accessory genomes of viruses reflect the integrated evolutionary history of the hosts they infected.
Keywords: E. coli; T4 phage; anti-phage defense; evolutionary biology; infectious disease; microbiology; phage counter-defense.
© 2022, Srikant, Guegler et al.
Conflict of interest statement
SS, CG No competing interests declared, ML Reviewing editor, eLife
Figures














References
-
- Appelmans R. Le dosage du bactériophage. Compt Rend Soc Biol. 1921;85:701
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

