Genic and chromosomal components of Prdm9-driven hybrid male sterility in mice (Mus musculus)
- PMID: 35924978
- PMCID: PMC9434306
- DOI: 10.1093/genetics/iyac116
Genic and chromosomal components of Prdm9-driven hybrid male sterility in mice (Mus musculus)
Abstract
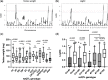
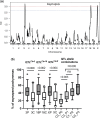
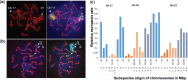
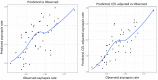
Hybrid sterility contributes to speciation by preventing gene flow between related taxa. Prdm9, the first and only hybrid male sterility gene known in vertebrates, predetermines the sites of recombination between homologous chromosomes and their synapsis in early meiotic prophase. The asymmetric binding of PRDM9 to heterosubspecific homologs of Mus musculus musculus × Mus musculus domesticus F1 hybrids and increase of PRDM9-independent DNA double-strand break hotspots results indificult- to- repair double-strand breaks, incomplete synapsis of homologous chromosomes, and meiotic arrest at the first meiotic prophase. Here, we show that Prdm9 behaves as a major hybrid male sterility gene in mice outside the Mus musculus musculus × Mus musculus domesticus F1 hybrids, in the genomes composed of Mus musculus castaneus and Mus musculus musculus chromosomes segregating on the Mus musculus domesticus background. The Prdm9cst/dom2 (castaneus/domesticus) allelic combination secures meiotic synapsis, testes weight, and sperm count within physiological limits, while the Prdm9msc1/dom2 (musculus/domesticus) males show a range of fertility impairment. Out of 5 quantitative trait loci contributing to the Prdm9msc1/dom2-related infertility, 4 control either meiotic synapsis or fertility phenotypes and 1 controls both, synapsis, and fertility. Whole-genome genotyping of individual chromosomes showed preferential involvement of nonrecombinant musculus chromosomes in asynapsis in accordance with the chromosomal character of hybrid male sterility. Moreover, we show that the overall asynapsis rate can be estimated solely from the genotype of individual males by scoring the effect of nonrecombinant musculus chromosomes. Prdm9-controlled hybrid male sterility represents an example of genetic architecture of hybrid male sterility consisting of genic and chromosomal components.
Keywords: HORMAD2; SYCP3; homologous synapsis; meiosis; spermatogenesis; synaptonemal complex.
© The Author(s) 2022. Published by Oxford University Press on behalf of Genetics Society of America. All rights reserved. For permissions, please email: journals.permissions@oup.com.
Figures


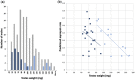
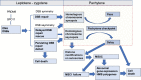
References
-
- Abe K, Noguchi H, Tagawa K, Yuzuriha M, Toyoda A, Kojima T, Ezawa K, Saitou N, Hattori M, Sakaki Y, et al Contribution of Asian mouse subspecies Mus musculus molossinus to genomic constitution of strain C57BL/6J, as defined by BAC-end sequence-SNP analysis. Genome Res. 2004;14(12):2439–2447. - PMC - PubMed
-
- Baird SJE, Macholan M, , , , .. What can the Mus musculus musculus/M. m. domesticus hybrid zone tell us about speciation? In: Macholan M, Baird SJ, Muclinger P, Pialek J (editors). Evolution of the House Mouse. Cambridge: Cambridge University Press; 2012. p. 334–372.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

