Evolution and molecular interactions of major histocompatibility complex (MHC)-G, -E and -F genes
- PMID: 35925520
- PMCID: PMC9352621
- DOI: 10.1007/s00018-022-04491-z
Evolution and molecular interactions of major histocompatibility complex (MHC)-G, -E and -F genes
Abstract
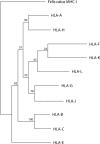
Classical HLA (Human Leukocyte Antigen) is the Major Histocompatibility Complex (MHC) in man. HLA genes and disease association has been studied at least since 1967 and no firm pathogenic mechanisms have been established yet. HLA-G immune modulation gene (and also -E and -F) are starting the same arduous way: statistics and allele association are the trending subjects with the same few results obtained by HLA classical genes, i.e., no pathogenesis may be discovered after many years of a great amount of researchers' effort. Thus, we believe that it is necessary to follow different research methodologies: (1) to approach this problem, based on how evolution has worked maintaining together a cluster of immune-related genes (the MHC) in a relatively short chromosome area since amniotes to human at least, i.e., immune regulatory genes (MHC-G, -E and -F), adaptive immune classical class I and II genes, non-adaptive immune genes like (C2, C4 and Bf) (2); in addition to using new in vitro models which explain pathogenetics of HLA and disease associations. In fact, this evolution may be quite reliably studied during about 40 million years by analyzing the evolution of MHC-G, -E, -F, and their receptors (KIR-killer-cell immunoglobulin-like receptor, NKG2-natural killer group 2-, or TCR-T-cell receptor-among others) in the primate evolutionary lineage, where orthology of these molecules is apparently established, although cladistic studies show that MHC-G and MHC-B genes are the ancestral class I genes, and that New World apes MHC-G is paralogous and not orthologous to all other apes and man MHC-G genes. In the present review, we outline past and possible future research topics: co-evolution of adaptive MHC classical (class I and II), non-adaptive (i.e., complement) and modulation (i.e., non-classical class I) immune genes may imply that the study of full or part of MHC haplotypes involving several loci/alleles instead of single alleles is important for uncovering HLA and disease pathogenesis. It would mainly apply to starting research on HLA-G extended haplotypes and disease association and not only using single HLA-G genetic markers.
Keywords: Apes; Complotypes; Disease; Evolution; HLA; HLA-E; HLA-F; HLA-G; Haplotypes; MHC; Monkeys.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no relevant competing interests to disclose.
Figures
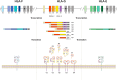

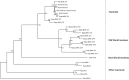
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

