Bioactive potentiality of secondary metabolites from endophytic bacteria against SARS-COV-2: An in-silico approach
- PMID: 35925905
- PMCID: PMC9352062
- DOI: 10.1371/journal.pone.0269962
Bioactive potentiality of secondary metabolites from endophytic bacteria against SARS-COV-2: An in-silico approach
Abstract
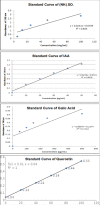
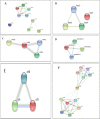
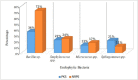
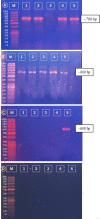
Five endophytic bacterial isolates were studied to identify morphologically and biochemically, according to established protocols and further confirmed by 16S rDNA Sanger sequencing, as Priestia megaterium, Staphylococcus caprae, Neobacillus drentensis, Micrococcus yunnanensis, and Sphingomonas paucimobiliz, which were then tested for phytohormone, ammonia, and hydrolytic enzyme production. Antioxidant compounds total phenolic content (TPC), and total flavonoid content (TFC) were assessed by using bacterial crude extracts obtained from 24-hour shake-flask culture. Phylogenetic tree analysis of those identified isolates shared sequence similarities with the members of Bacillus, Micrococcus, Staphylococcus, and Pseudomonas species, and after GenBank submission, accession numbers for the nucleotide sequences were found to be MW494406, MW494408, MW494401, MW494402, and MZ021340, respectively. In silico analysis was performed to identify their bioactive genes and compounds in the context of bioactive secondary metabolite production with medicinal value, where nine significant bioactive compounds according to six different types of bioactive secondary metabolites were identified, and their structures, gene associations, and protein-protein networks were analyzed by different computational tools and servers, which were reported earlier with their antimicrobial, anti-infective, antioxidant, and anti-cancer capabilities. These compounds were then docked to the 3-chymotrypsin-like protease (3CLpro) of the novel SARS-COV-2. Docking scores were then compared with 3CLpro reference inhibitor (lopinavir), and docked compounds were further subjected to ADMET and drug-likeness analyses. Ligand-protein interactions showed that two compounds (microansamycin and aureusimine) interacted favorably with coronavirus 3CLpro. Besides, in silico analysis, we also performed NMR for metabolite detection whereas three metabolites (microansamycin, aureusimine, and stenothricin) were confirmed from the 1H NMR profiles. As a consequence, the metabolites found from NMR data aligned with our in-silico analysis that carries a significant outcome of this research. Finally, Endophytic bacteria collected from medicinal plants can provide new leading bioactive compounds against target proteins of SARS-COV-2, which could be an effective approach to accelerate drug innovation and development.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures









References
-
- Schulz B. Mutualistic interactions with fungal root endophytes. Microbial root endophytes. Springer; 2006. pp. 261–279.
-
- Wilson D. Endophyte: the evolution of a term, and clarification of its use and definition. Oikos. 1995; 274–276.
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

