Multi-label classification of symptom terms from free-text bilingual adverse drug reaction reports using natural language processing
- PMID: 35925971
- PMCID: PMC9352066
- DOI: 10.1371/journal.pone.0270595
Multi-label classification of symptom terms from free-text bilingual adverse drug reaction reports using natural language processing
Abstract
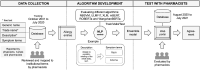
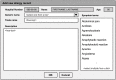
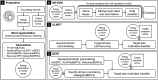
Allergic reactions to medication range from mild to severe or even life-threatening. Proper documentation of patient allergy information is critical for safe prescription, avoiding drug interactions, and reducing healthcare costs. Allergy information is regularly obtained during the medical interview, but is often poorly documented in electronic health records (EHRs). While many EHRs allow for structured adverse drug reaction (ADR) reporting, a free-text entry is still common. The resulting information is neither interoperable nor easily reusable for other applications, such as clinical decision support systems and prescription alerts. Current approaches require pharmacists to review and code ADRs documented by healthcare professionals. Recently, the effectiveness of machine algorithms in natural language processing (NLP) has been widely demonstrated. Our study aims to develop and evaluate different NLP algorithms that can encode unstructured ADRs stored in EHRs into institutional symptom terms. Our dataset consists of 79,712 pharmacist-reviewed drug allergy records. We evaluated three NLP techniques: Naive Bayes-Support Vector Machine (NB-SVM), Universal Language Model Fine-tuning (ULMFiT), and Bidirectional Encoder Representations from Transformers (BERT). We tested different general-domain pre-trained BERT models, including mBERT, XLM-RoBERTa, and WanchanBERTa, as well as our domain-specific AllergyRoBERTa, which was pre-trained from scratch on our corpus. Overall, BERT models had the highest performance. NB-SVM outperformed ULMFiT and BERT for several symptom terms that are not frequently coded. The ensemble model achieved an exact match ratio of 95.33%, a F1 score of 98.88%, and a mean average precision of 97.07% for the 36 most frequently coded symptom terms. The model was then further developed into a symptom term suggestion system and achieved a Krippendorff's alpha agreement coefficient of 0.7081 in prospective testing with pharmacists. Some degree of automation could both accelerate the availability of allergy information and reduce the efforts for human coding.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
Similar articles
-
A Natural Language Processing Model for COVID-19 Detection Based on Dutch General Practice Electronic Health Records by Using Bidirectional Encoder Representations From Transformers: Development and Validation Study.J Med Internet Res. 2023 Oct 4;25:e49944. doi: 10.2196/49944. J Med Internet Res. 2023. PMID: 37792444 Free PMC article.
-
Automation of penicillin adverse drug reaction categorisation and risk stratification with machine learning natural language processing.Int J Med Inform. 2021 Dec;156:104611. doi: 10.1016/j.ijmedinf.2021.104611. Epub 2021 Oct 5. Int J Med Inform. 2021. PMID: 34653809
-
Evaluation of a BERT Natural Language Processing Model for Automating CT and MRI Triage and Protocol Selection.Can Assoc Radiol J. 2025 May;76(2):265-272. doi: 10.1177/08465371241255895. Epub 2024 Jun 4. Can Assoc Radiol J. 2025. PMID: 38832645
-
Natural language processing of symptoms documented in free-text narratives of electronic health records: a systematic review.J Am Med Inform Assoc. 2019 Apr 1;26(4):364-379. doi: 10.1093/jamia/ocy173. J Am Med Inform Assoc. 2019. PMID: 30726935 Free PMC article.
-
A systematic review of natural language processing for classification tasks in the field of incident reporting and adverse event analysis.Int J Med Inform. 2019 Dec;132:103971. doi: 10.1016/j.ijmedinf.2019.103971. Epub 2019 Oct 5. Int J Med Inform. 2019. PMID: 31630063
Cited by
-
The Indonesian Young-Adult Attachment (IYAA): An audio-video dataset for behavioral young-adult attachment assessment.Data Brief. 2023 Sep 21;50:109599. doi: 10.1016/j.dib.2023.109599. eCollection 2023 Oct. Data Brief. 2023. PMID: 37780464 Free PMC article.
-
Application of the transformer model algorithm in chinese word sense disambiguation: a case study in chinese language.Sci Rep. 2024 Mar 15;14(1):6320. doi: 10.1038/s41598-024-56976-5. Sci Rep. 2024. PMID: 38491085 Free PMC article.
-
Optimizing classification of diseases through language model analysis of symptoms.Sci Rep. 2024 Jan 17;14(1):1507. doi: 10.1038/s41598-024-51615-5. Sci Rep. 2024. PMID: 38233458 Free PMC article.
-
Understanding patterns of loneliness in older long-term care users using natural language processing with free text case notes.PLoS One. 2025 Apr 2;20(4):e0319745. doi: 10.1371/journal.pone.0319745. eCollection 2025. PLoS One. 2025. PMID: 40173389 Free PMC article.
-
Construction of a Multi-Label Classifier for Extracting Multiple Incident Factors From Medication Incident Reports in Residential Care Facilities: Natural Language Processing Approach.JMIR Med Inform. 2024 Jul 23;12:e58141. doi: 10.2196/58141. JMIR Med Inform. 2024. PMID: 39042454 Free PMC article.
References
MeSH terms
LinkOut - more resources
Full Text Sources
Medical
Research Materials

