Human UFSP1 is an active protease that regulates UFM1 maturation and UFMylation
- PMID: 35926457
- PMCID: PMC9638016
- DOI: 10.1016/j.celrep.2022.111168
Human UFSP1 is an active protease that regulates UFM1 maturation and UFMylation
Abstract
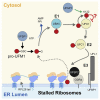
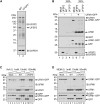
An essential first step in the post-translational modification of proteins with UFM1, UFMylation, is the proteolytic cleavage of pro-UFM1 to expose a C-terminal glycine. Of the two UFM1-specific proteases (UFSPs) identified in humans, only UFSP2 is reported to be active, since the annotated sequence of UFSP1 lacks critical catalytic residues. Nonetheless, efficient UFM1 maturation occurs in cells lacking UFSP2, suggesting the presence of another active protease. We herein identify UFSP1 translated from a non-canonical start site to be this protease. Cells lacking both UFSPs show complete loss of UFMylation resulting from an absence of mature UFM1. While UFSP2, but not UFSP1, removes UFM1 from the ribosomal subunit RPL26, UFSP1 acts earlier in the pathway to mature UFM1 and cleave a potential autoinhibitory modification on UFC1, thereby controlling activation of UFMylation. In summary, our studies reveal important distinctions in substrate specificity and localization-dependent functions for the two proteases in regulating UFMylation.
Keywords: CP: Cell biology; CP: Molecular biology; ER; UBA5; UFC1; UFM1; cysteine protease; endoplasmic reticulum; membrane protein; ribosome; ubiquitin; ubiquitin-like modifier.
Copyright © 2022 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures






Similar articles
-
Human UFSP1 translated from an upstream near-cognate initiation codon functions as an active UFM1-specific protease.J Biol Chem. 2022 Jun;298(6):102016. doi: 10.1016/j.jbc.2022.102016. Epub 2022 May 5. J Biol Chem. 2022. PMID: 35525273 Free PMC article.
-
Nontraditional translation is the key to UFMylation and beyond.J Biol Chem. 2022 Oct;298(10):102431. doi: 10.1016/j.jbc.2022.102431. Epub 2022 Aug 28. J Biol Chem. 2022. PMID: 36037969 Free PMC article.
-
Two novel ubiquitin-fold modifier 1 (Ufm1)-specific proteases, UfSP1 and UfSP2.J Biol Chem. 2007 Feb 23;282(8):5256-62. doi: 10.1074/jbc.M610590200. Epub 2006 Dec 20. J Biol Chem. 2007. PMID: 17182609
-
UFMylation: A Unique & Fashionable Modification for Life.Genomics Proteomics Bioinformatics. 2016 Jun;14(3):140-146. doi: 10.1016/j.gpb.2016.04.001. Epub 2016 May 20. Genomics Proteomics Bioinformatics. 2016. PMID: 27212118 Free PMC article. Review.
-
Emerging role of protein modification by UFM1 in cancer.Biochem Biophys Res Commun. 2022 Dec 10;633:61-63. doi: 10.1016/j.bbrc.2022.08.093. Biochem Biophys Res Commun. 2022. PMID: 36344165 Review.
Cited by
-
The UFM1 system regulates ER-phagy through the ufmylation of CYB5R3.Nat Commun. 2022 Dec 21;13(1):7857. doi: 10.1038/s41467-022-35501-0. Nat Commun. 2022. PMID: 36543799 Free PMC article.
-
The UFMylation pathway is impaired in Alzheimer's disease.Mol Neurodegener. 2024 Dec 18;19(1):97. doi: 10.1186/s13024-024-00784-y. Mol Neurodegener. 2024. PMID: 39696466 Free PMC article.
-
Multifaceted roles of UFMylation in health and disease.Acta Pharmacol Sin. 2025 Apr;46(4):805-815. doi: 10.1038/s41401-024-01456-9. Epub 2025 Jan 7. Acta Pharmacol Sin. 2025. PMID: 39775503 Review.
-
Mono-UFMylation promotes misfolding-associated secretion of α-synuclein.Sci Adv. 2024 Mar 15;10(11):eadk2542. doi: 10.1126/sciadv.adk2542. Epub 2024 Mar 15. Sci Adv. 2024. PMID: 38489364 Free PMC article.
-
The ufmylation cascade controls COPII recruitment, anterograde transport, and sorting of nascent GPCRs at ER.Sci Adv. 2024 Jun 21;10(25):eadm9216. doi: 10.1126/sciadv.adm9216. Epub 2024 Jun 21. Sci Adv. 2024. PMID: 38905340 Free PMC article.
References
-
- Colin E., Daniel J., Ziegler A., Wakim J., Scrivo A., Haack T.B., Khiati S., Denommé A.S., Amati-Bonneau P., Charif M., et al. Biallelic variants in UBA5 reveal that disruption of the UFM1 cascade can result in early-onset encephalopathy. Am. J. Hum. Genet. 2016;99:695–703. doi: 10.1016/j.ajhg.2016.06.030. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

