Interactions between mTORC2 core subunits Rictor and mSin1 dictate selective and context-dependent phosphorylation of substrate kinases SGK1 and Akt
- PMID: 35926713
- PMCID: PMC9440446
- DOI: 10.1016/j.jbc.2022.102288
Interactions between mTORC2 core subunits Rictor and mSin1 dictate selective and context-dependent phosphorylation of substrate kinases SGK1 and Akt
Abstract
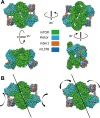
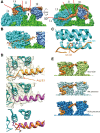
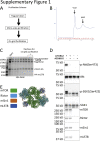
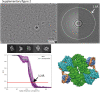
Mechanistic target of rapamycin complex 2 (mTORC2) is a multi-subunit kinase complex, central to multiple essential signaling pathways. Two core subunits, Rictor and mSin1, distinguish it from the related mTORC1 and support context-dependent phosphorylation of its substrates. mTORC2 structures have been determined previously; however, important questions remain, particularly regarding the structural determinants mediating substrate specificity and context-dependent activity. Here, we used cryo-EM to obtain high-resolution structures of the human mTORC2 apo-complex in the presence of substrates Akt and SGK1. Using functional assays, we then tested predictions suggested by substrate-induced structural changes in mTORC2. For the first time, we visualized in the apo-state the side chain interactions between Rictor and mTOR that sterically occlude recruitment of mTORC1 substrates and confer resistance to the mTORC1 inhibitor rapamycin. Also in the apo-state, we observed that mSin1 formed extensive contacts with Rictor via a pair of short α-helices nestled between two Rictor helical repeat clusters, as well as by an extended strand that makes multiple weak contacts with Rictor helical cluster 1. In co-complex structures, we found that SGK1, but not Akt, markedly altered the conformation of the mSin1 N-terminal extended strand, disrupting multiple weak interactions while inducing a large rotation of mSin1 residue Arg-83, which then interacts with a patch of negatively charged residues within Rictor. Finally, we demonstrate mutation of Arg-83 to Ala selectively disrupts mTORC2-dependent phosphorylation of SGK1, but not of Akt, supporting context-dependent substrate selection. These findings provide new structural and functional insights into mTORC2 specificity and context-dependent activity.
Keywords: Akt; SGK1; conformation change; cryo-EM; mTORC2; structure; substrate specificity.
Copyright © 2022 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Conflict of interest Y. C. is an Investigator of Howard Hughes Medical Institute. All other authors claim no conflict of interest.
Figures





References
-
- Heitman J., Movva N.R., Hall M.N. Targets for cell cycle arrest by the immunosuppressant rapamycin in yeast. Science. 1991;253:905–909. - PubMed
-
- Sabatini D.M., Erdjument-Bromage H., Lui M., Tempst P., Snyder S.H. RAFT1: a mammalian protein that binds to FKBP12 in a rapamycin-dependent fashion and is homologous to yeast TORs. Cell. 1994;78:35–43. - PubMed
-
- Saxton R.A., Sabatini D.M. mTOR signaling in growth, metabolism, and disease. Cell. 2017;169:361–371. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

