Ubiquitome profiling reveals a regulatory pattern of UPL3 with UBP12 on metabolic-leaf senescence
- PMID: 35926874
- PMCID: PMC9354775
- DOI: 10.26508/lsa.202201492
Ubiquitome profiling reveals a regulatory pattern of UPL3 with UBP12 on metabolic-leaf senescence
Abstract
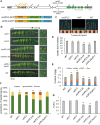
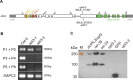
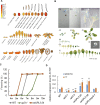
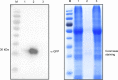
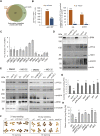
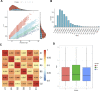
The HECT-type UPL3 ligase plays critical roles in plant development and stress protection, but understanding of its regulation remains limited. Here, the multi-omics analyses of ubiquitinated proteins in <i>upl3</i> mutants were performed. A landscape of UPL3-dependent ubiquitinated proteins is constructed: Preferential ubiquitination of proteins related to carbon fixation represented the largest set of proteins with increased ubiquitination in the <i>upl3</i> plant, including most of carbohydrate metabolic enzymes, BRM, and variant histone, whereas a small set of proteins with reduced ubiquitination caused by the <i>upl3</i> mutation were linked to cysteine/methionine synthesis, as well as hexokinase 1 (HXK1) and phosphoenolpyruvate carboxylase 2 (PPC2). Notably, ubiquitin hydrolase 12 (UBP12), BRM, HXK1, and PPC2 were identified as the UPL3-interacting partners in vivo and in vitro. Characterization of <i>brm</i>, <i>upl3</i>, <i>ppc2</i>, <i>gin2</i>, and <i>ubp12</i> mutant plants and proteomic and transcriptomic analysis suggested that UPL3 fine-tunes carbohydrate metabolism, mediating cellular senescence by interacting with UBP12, BRM, HXK1, and PPC2. Our results highlight a regulatory pattern of UPL3 with UBP12 as a hub of regulator on proteolysis-independent regulation and proteolysis-dependent degradation.
© 2022 Lan et al.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures















Similar articles
-
The HECT ubiquitin-protein ligase (UPL) family in Arabidopsis: UPL3 has a specific role in trichome development.Plant J. 2003 Sep;35(6):729-42. doi: 10.1046/j.1365-313x.2003.01844.x. Plant J. 2003. PMID: 12969426
-
Proteasome-associated HECT-type ubiquitin ligase activity is required for plant immunity.PLoS Pathog. 2018 Nov 20;14(11):e1007447. doi: 10.1371/journal.ppat.1007447. eCollection 2018 Nov. PLoS Pathog. 2018. PMID: 30458055 Free PMC article.
-
Arabidopsis ubiquitin-specific proteases UBP12 and UBP13 shape ORE1 levels during leaf senescence induced by nitrogen deficiency.New Phytol. 2019 Aug;223(3):1447-1460. doi: 10.1111/nph.15879. Epub 2019 Jun 17. New Phytol. 2019. PMID: 31050353
-
Insights into a functional model of key deubiquitinases UBP12/13 in plants.New Phytol. 2024 Apr;242(2):424-430. doi: 10.1111/nph.19639. Epub 2024 Feb 26. New Phytol. 2024. PMID: 38406992 Review.
-
Leaf senescence and abiotic stresses share reactive oxygen species-mediated chloroplast degradation.Protoplasma. 2012 Jul;249(3):469-81. doi: 10.1007/s00709-011-0308-z. Epub 2011 Jul 31. Protoplasma. 2012. PMID: 21805384 Review.
Cited by
-
UPL3 promotes BZR1 degradation, growth arrest, and seedling survival under starvation stress in Arabidopsis.Plant Commun. 2025 Jul 14;6(7):101389. doi: 10.1016/j.xplc.2025.101389. Epub 2025 May 28. Plant Commun. 2025. PMID: 40443036 Free PMC article.
-
Comparative proteomics in tall fescue to reveal underlying mechanisms for improving Photosystem II thermotolerance during heat stress memory.BMC Genomics. 2024 Jul 9;25(1):683. doi: 10.1186/s12864-024-10580-z. BMC Genomics. 2024. PMID: 38982385 Free PMC article.
-
Epigenetic control of plant senescence and cell death and its application in crop improvement.Front Plant Sci. 2023 Oct 30;14:1258487. doi: 10.3389/fpls.2023.1258487. eCollection 2023. Front Plant Sci. 2023. PMID: 37965008 Free PMC article. Review.
-
An insight into the roles of ubiquitin-specific proteases in plants: development and growth, morphogenesis, and stress response.Front Plant Sci. 2024 Jun 27;15:1396634. doi: 10.3389/fpls.2024.1396634. eCollection 2024. Front Plant Sci. 2024. PMID: 38993940 Free PMC article. Review.
References
-
- Archacki R, Yatusevich R, Buszewicz D, Krzyczmonik K, Patryn J, Iwanicka-Nowicka R, Biecek P, Wilczynski B, Koblowska M, Jerzmanowski A, et al. (2017) Arabidopsis SWI/SNF chromatin remodeling complex binds both promoters and terminators to regulate gene expression. Nucleic Acids Res 45: 3116–3129. 10.1093/nar/gkw1273 - DOI - PMC - PubMed
-
- Bensussan M, Lefebvre V, Ducamp A, Trouverie J, Gineau E, Fortabat MN, Guillebaux A, Baldy A, Naquin D, Herbette S, et al. (2015) Suppression of dwarf and irregular xylem phenotypes generates low-acetylated biomass lines in Arabidopsis. Plant Physiol 168: 452–463. 10.1104/pp.15.00122 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous
