Simultaneous monitoring of eight human respiratory viruses including SARS-CoV-2 using liquid chromatography-tandem mass spectrometry
- PMID: 35927299
- PMCID: PMC9352774
- DOI: 10.1038/s41598-022-16250-y
Simultaneous monitoring of eight human respiratory viruses including SARS-CoV-2 using liquid chromatography-tandem mass spectrometry
Abstract
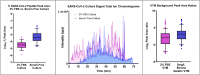
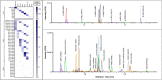
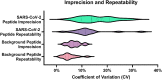
Diagnosis of Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) infection has primarily been achieved using reverse transcriptase polymerase chain reaction (RT-PCR) for acute infection, and serology for prior infection. Assay with RT-PCR provides data on presence or absence of viral RNA, with no information on virus replication competence, infectivity, or virus characterisation. Liquid chromatography-tandem mass spectrometry (LC-MS/MS) is typically not used in clinical virology, despite its potential to provide supplemental data about the presence of viral proteins and thus the potential for replication-competent, transmissible virus. Using the SARS-CoV-2 as a model virus, we developed a fast 'bottom-up' proteomics workflow for discovery of target virus peptides using 'serum-free' culture conditions, providing high coverage of viral proteins without the need for protein or peptide fractionation techniques. This workflow was then applied to Coronaviruses OC43 and 229E, Influenza A/H1N1 and H3N2, Influenza B, and Respiratory Syncytial Viruses A and B. Finally, we created an LC-MS/MS method for targeted detection of the eight-virus panel in clinical specimens, successfully detecting peptides from the SARS-CoV-2 ORF9B and nucleoprotein in RT-PCR positive samples. The method provides specific detection of respiratory viruses from clinical samples containing moderate viral loads and is an important further step to the use of LC-MS/MS in diagnosis of viral infection.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures


Similar articles
-
Evaluation of Three Multiplex Real-time Reverse Transcription PCR Assays for Simultaneous Detection of SARS-CoV-2, Influenza A/B, and Respiratory Syncytial Virus in Nasopharyngeal Swabs.J Korean Med Sci. 2021 Dec 13;36(48):e328. doi: 10.3346/jkms.2021.36.e328. J Korean Med Sci. 2021. PMID: 34904407 Free PMC article.
-
Developing a Loop-Mediated Isothermal Amplification Assay for the Rapid Detection of Seven Respiratory Viruses including SARS-CoV-2.Medicina (Kaunas). 2022 Sep 5;58(9):1224. doi: 10.3390/medicina58091224. Medicina (Kaunas). 2022. PMID: 36143901 Free PMC article.
-
Novel dual multiplex real-time RT-PCR assays for the rapid detection of SARS-CoV-2, influenza A/B, and respiratory syncytial virus using the BD MAX open system.Emerg Microbes Infect. 2021 Dec;10(1):161-166. doi: 10.1080/22221751.2021.1873073. Emerg Microbes Infect. 2021. PMID: 33410371 Free PMC article.
-
[Applications of separation technology in novel coronavirus research, epidemic prevention and detection].Se Pu. 2021 Jul 8;39(7):679-685. doi: 10.3724/SP.J.1123.2021.03022. Se Pu. 2021. PMID: 34227364 Free PMC article. Review. Chinese.
-
High resolution mass spectrometry of respiratory viruses: beyond MALDI-ToF instruments for next generation viral typing, subtyping, variant and sub-variant identification.Analyst. 2023 Sep 11;148(18):4263-4273. doi: 10.1039/d3an00953j. Analyst. 2023. PMID: 37587867 Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

