The ribosome stabilizes partially folded intermediates of a nascent multi-domain protein
- PMID: 35927328
- PMCID: PMC7613651
- DOI: 10.1038/s41557-022-01004-0
The ribosome stabilizes partially folded intermediates of a nascent multi-domain protein
Abstract
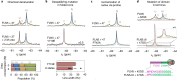
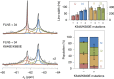
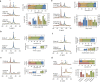
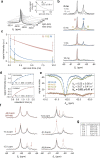
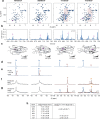
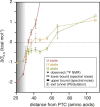
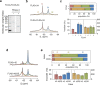
Co-translational folding is crucial to ensure the production of biologically active proteins. The ribosome can alter the folding pathways of nascent polypeptide chains, yet a structural understanding remains largely inaccessible experimentally. We have developed site-specific labelling of nascent chains to detect and measure, using 19F nuclear magnetic resonance (NMR) spectroscopy, multiple states accessed by an immunoglobulin-like domain within a tandem repeat protein during biosynthesis. By examining ribosomes arrested at different stages during translation of this common structural motif, we observe highly broadened NMR resonances attributable to two previously unidentified intermediates, which are stably populated across a wide folding transition. Using molecular dynamics simulations and corroborated by cryo-electron microscopy, we obtain models of these partially folded states, enabling experimental verification of a ribosome-binding site that contributes to their high stabilities. We thus demonstrate a mechanism by which the ribosome could thermodynamically regulate folding and other co-translational processes.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures


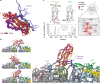






References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

