Sample-multiplexing approaches for single-cell sequencing
- PMID: 35927335
- PMCID: PMC11073057
- DOI: 10.1007/s00018-022-04482-0
Sample-multiplexing approaches for single-cell sequencing
Abstract
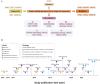
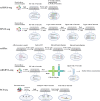
Single-cell sequencing is widely used in biological and medical studies. However, its application with multiple samples is hindered by inefficient sample processing, high experimental costs, ambiguous identification of true single cells, and technical batch effects. Here, we introduce sample-multiplexing approaches for single-cell sequencing in transcriptomics, epigenomics, genomics, and multiomics. In single-cell transcriptomics, sample multiplexing uses variants of native or artificial features as sample markers, enabling sample pooling and decoding. Such features include: (1) natural genetic variation, (2) nucleotide-barcode anchoring on cellular or nuclear membranes, (3) nucleotide-barcode internalization to the cytoplasm or nucleus, (4) vector-based barcode expression in cells, and (5) nucleotide-barcode incorporation during library construction. Other single-cell omics methods are based on similar concepts, particularly single-cell combinatorial indexing. These methods overcome current challenges, while enabling super-loading of single cells. Finally, selection guidelines are presented that can accelerate technological application.
Keywords: Cell Hashing; Multi-omics; Spatial transcriptomics; scATAC-seq; scRNA-seq.
© 2022. The Author(s), under exclusive licence to Springer Nature Switzerland AG.
Conflict of interest statement
There are no conflicts of interest to declare.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
- SZBL2020090501003/Open Fund Programs of Shenzhen Bay Laboratory
- Major Basic Cultivation Project 2018B030308004/Natural Science Foundation of Guangdong Province
- Major Projects of Basic/Natural Science Foundation of Guangdong Province
- Applied Basic Research 2019B1515120033/Natural Science Foundation of Guangdong Province
- 32071452/National Natural Science Foundation of China
LinkOut - more resources
Full Text Sources

