Nucleotide imbalance decouples cell growth from cell proliferation
- PMID: 35927450
- PMCID: PMC9359916
- DOI: 10.1038/s41556-022-00965-1
Nucleotide imbalance decouples cell growth from cell proliferation
Abstract
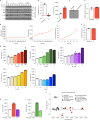
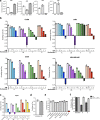
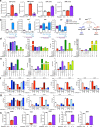
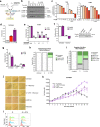
Nucleotide metabolism supports RNA synthesis and DNA replication to enable cell growth and division. Nucleotide depletion can inhibit cell growth and proliferation, but how cells sense and respond to changes in the relative levels of individual nucleotides is unclear. Moreover, the nucleotide requirement for biomass production changes over the course of the cell cycle, and how cells coordinate differential nucleotide demands with cell cycle progression is not well understood. Here we find that excess levels of individual nucleotides can inhibit proliferation by disrupting the relative levels of nucleotide bases needed for DNA replication and impeding DNA replication. The resulting purine and pyrimidine imbalances are not sensed by canonical growth regulatory pathways like mTORC1, Akt and AMPK signalling cascades, causing excessive cell growth despite inhibited proliferation. Instead, cells rely on replication stress signalling to survive during, and recover from, nucleotide imbalance during S phase. We find that ATR-dependent replication stress signalling is activated during unperturbed S phases and promotes nucleotide availability to support DNA replication. Together, these data reveal that imbalanced nucleotide levels are not detected until S phase, rendering cells reliant on replication stress signalling to cope with this metabolic problem and disrupting the coordination of cell growth and division.
© 2022. The Author(s).
Conflict of interest statement
The authors are aware of no direct conflicts with the topic of the study; however, F.F.D. and M.G.V.H. are included on a patent application regarding impact of nucleotide imbalance on cell state. S.R.M. declares he is a co-founder of Affinity Biosensors and Travera, and M.G.V.H. declares he is an advisory board member for Agios Pharmaceuticals, Aeglea Biotherapeutics, Faeth Therapeutics, Drioa Ventures and iTeos Therapeutics, and a co-founder of Auron Therapeutics. The other authors declare no competing interests.
Figures









Similar articles
-
mTORC1 Couples Nucleotide Synthesis to Nucleotide Demand Resulting in a Targetable Metabolic Vulnerability.Cancer Cell. 2017 Nov 13;32(5):624-638.e5. doi: 10.1016/j.ccell.2017.09.013. Epub 2017 Oct 19. Cancer Cell. 2017. PMID: 29056426 Free PMC article.
-
Differential control of cell cycle, proliferation, and survival of primary T lymphocytes by purine and pyrimidine nucleotides.J Immunol. 2003 May 15;170(10):4986-95. doi: 10.4049/jimmunol.170.10.4986. J Immunol. 2003. PMID: 12734342
-
CD98hc (SLC3A2) sustains amino acid and nucleotide availability for cell cycle progression.Sci Rep. 2019 Oct 1;9(1):14065. doi: 10.1038/s41598-019-50547-9. Sci Rep. 2019. PMID: 31575908 Free PMC article.
-
The Contribution of Lysosomes to DNA Replication.Cells. 2021 Apr 30;10(5):1068. doi: 10.3390/cells10051068. Cells. 2021. PMID: 33946407 Free PMC article. Review.
-
Regulation of Cell Division in Bacteria by Monitoring Genome Integrity and DNA Replication Status.J Bacteriol. 2020 Jan 2;202(2):e00408-19. doi: 10.1128/JB.00408-19. Print 2020 Jan 2. J Bacteriol. 2020. PMID: 31548275 Free PMC article. Review.
Cited by
-
RNASET2 Deficiency Induces Hepatocellular Carcinoma Metastasis through Cholesterol-Triggered MET Activation.Adv Sci (Weinh). 2025 Mar;12(12):e2411888. doi: 10.1002/advs.202411888. Epub 2025 Feb 4. Adv Sci (Weinh). 2025. PMID: 39903758 Free PMC article.
-
Human cytomegalovirus UL82 promotes cell cycle progression of colorectal cancer by upregulating AGR2.Commun Biol. 2025 Feb 17;8(1):251. doi: 10.1038/s42003-025-07674-z. Commun Biol. 2025. PMID: 39962326 Free PMC article.
-
De novo and salvage purine synthesis pathways across tissues and tumors.Cell. 2024 Jul 11;187(14):3602-3618.e20. doi: 10.1016/j.cell.2024.05.011. Epub 2024 May 31. Cell. 2024. PMID: 38823389 Free PMC article.
-
Allopurinol treatment changes microglial characteristics in neonatal mice.MicroPubl Biol. 2025 Jul 30;2025:10.17912/micropub.biology.001739. doi: 10.17912/micropub.biology.001739. eCollection 2025. MicroPubl Biol. 2025. PMID: 40838120 Free PMC article.
-
Intrinsic PARG inhibitor sensitivity is mimicked by TIMELESS haploinsufficiency and rescued by nucleoside supplementation.NAR Cancer. 2024 Jul 16;6(3):zcae030. doi: 10.1093/narcan/zcae030. eCollection 2024 Sep. NAR Cancer. 2024. PMID: 39015544 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

