PROBER identifies proteins associated with programmable sequence-specific DNA in living cells
- PMID: 35927480
- PMCID: PMC10202087
- DOI: 10.1038/s41592-022-01552-w
PROBER identifies proteins associated with programmable sequence-specific DNA in living cells
Abstract
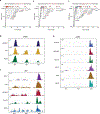
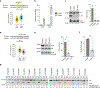
DNA-protein interactions mediate physiologic gene regulation and may be altered by DNA variants linked to polygenic disease. To enhance the speed and signal-to-noise ratio (SNR) in the identification and quantification of proteins associated with specific DNA sequences in living cells, we developed proximal biotinylation by episomal recruitment (PROBER). PROBER uses high-copy episomes to amplify SNR, and proximity proteomics (BioID) to identify the transcription factors and additional gene regulators associated with short DNA sequences of interest. PROBER quantified both constitutive and inducible association of transcription factors and corresponding chromatin regulators to target DNA sequences and binding quantitative trait loci due to single-nucleotide variants. PROBER identified alterations in regulator associations due to cancer hotspot mutations in the hTERT promoter, indicating that these mutations increase promoter association with specific gene activators. PROBER provides an approach to rapidly identify proteins associated with specific DNA sequences and their variants in living cells.
© 2022. This is a U.S. Government work and not under copyright protection in the US; foreign copyright protection may apply.
Conflict of interest statement
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
Figures






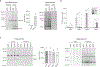






Similar articles
-
BioID screening of biotinylation sites using the avidin-like protein Tamavidin 2-REV identifies global interactors of stimulator of interferon genes (STING).J Biol Chem. 2020 Aug 7;295(32):11174-11183. doi: 10.1074/jbc.RA120.014323. Epub 2020 Jun 17. J Biol Chem. 2020. PMID: 32554809 Free PMC article.
-
Proximity biotinylation and affinity purification are complementary approaches for the interactome mapping of chromatin-associated protein complexes.J Proteomics. 2015 Apr 6;118:81-94. doi: 10.1016/j.jprot.2014.09.011. Epub 2014 Oct 2. J Proteomics. 2015. PMID: 25281560 Free PMC article.
-
BioID identifies novel c-MYC interacting partners in cultured cells and xenograft tumors.J Proteomics. 2015 Apr 6;118:95-111. doi: 10.1016/j.jprot.2014.09.029. Epub 2014 Oct 18. J Proteomics. 2015. PMID: 25452129
-
Changing the DNA landscape: putting a SPN on chromatin.Curr Top Microbiol Immunol. 2003;274:171-201. doi: 10.1007/978-3-642-55747-7_7. Curr Top Microbiol Immunol. 2003. PMID: 12596908 Review.
-
HMGI/Y proteins: flexible regulators of transcription and chromatin structure.Biochim Biophys Acta. 2001 May 28;1519(1-2):13-29. doi: 10.1016/s0167-4781(01)00215-9. Biochim Biophys Acta. 2001. PMID: 11406267 Review.
Cited by
-
In situ proximity labeling of proteins associated with circRNA and RNA G-quadruplexes.Nat Chem Biol. 2025 Aug 7. doi: 10.1038/s41589-025-01993-2. Online ahead of print. Nat Chem Biol. 2025. PMID: 40775047
-
Functional analysis of cancer-associated germline risk variants.Nat Genet. 2025 Mar;57(3):718-728. doi: 10.1038/s41588-024-02070-5. Epub 2025 Feb 17. Nat Genet. 2025. PMID: 39962238
-
Spatiotemporal and global profiling of DNA-protein interactions enables discovery of low-affinity transcription factors.Nat Chem. 2023 Jun;15(6):803-814. doi: 10.1038/s41557-023-01196-z. Epub 2023 Apr 27. Nat Chem. 2023. PMID: 37106095
-
Mass Spectrometry for Assessing Protein-Nucleic Acid Interactions.Anal Chem. 2023 Jan 10;95(1):115-127. doi: 10.1021/acs.analchem.2c04353. Anal Chem. 2023. PMID: 36625126 Free PMC article. Review. No abstract available.
-
Characterizing crosstalk in epigenetic signaling to understand disease physiology.Biochem J. 2023 Jan 13;480(1):57-85. doi: 10.1042/BCJ20220550. Biochem J. 2023. PMID: 36630129 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

