A deep learning framework for identifying essential proteins based on multiple biological information
- PMID: 35927611
- PMCID: PMC9351218
- DOI: 10.1186/s12859-022-04868-8
A deep learning framework for identifying essential proteins based on multiple biological information
Abstract
Background: Essential Proteins are demonstrated to exert vital functions on cellular processes and are indispensable for the survival and reproduction of the organism. Traditional centrality methods perform poorly on complex protein-protein interaction (PPI) networks. Machine learning approaches based on high-throughput data lack the exploitation of the temporal and spatial dimensions of biological information.
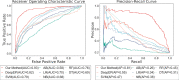
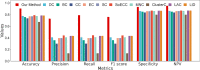
Results: We put forward a deep learning framework to predict essential proteins by integrating features obtained from the PPI network, subcellular localization, and gene expression profiles. In our model, the node2vec method is applied to learn continuous feature representations for proteins in the PPI network, which capture the diversity of connectivity patterns in the network. The concept of depthwise separable convolution is employed on gene expression profiles to extract properties and observe the trends of gene expression over time under different experimental conditions. Subcellular localization information is mapped into a long one-dimensional vector to capture its characteristics. Additionally, we use a sampling method to mitigate the impact of imbalanced learning when training the model. With experiments carried out on the data of Saccharomyces cerevisiae, results show that our model outperforms traditional centrality methods and machine learning methods. Likewise, the comparative experiments have manifested that our process of various biological information is preferable.
Conclusions: Our proposed deep learning framework effectively identifies essential proteins by integrating multiple biological data, proving a broader selection of subcellular localization information significantly improves the results of prediction and depthwise separable convolution implemented on gene expression profiles enhances the performance.
Keywords: Deep learning; Essential protein; Gene expression; Protein–protein interaction network; Subcellular localization.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no conflict of interest.
Figures











Similar articles
-
A Deep Learning Framework for Identifying Essential Proteins by Integrating Multiple Types of Biological Information.IEEE/ACM Trans Comput Biol Bioinform. 2021 Jan-Feb;18(1):296-305. doi: 10.1109/TCBB.2019.2897679. Epub 2021 Feb 3. IEEE/ACM Trans Comput Biol Bioinform. 2021. PMID: 30736002
-
DeepEP: a deep learning framework for identifying essential proteins.BMC Bioinformatics. 2019 Dec 2;20(Suppl 16):506. doi: 10.1186/s12859-019-3076-y. BMC Bioinformatics. 2019. PMID: 31787076 Free PMC article.
-
DeepHE: Accurately predicting human essential genes based on deep learning.PLoS Comput Biol. 2020 Sep 16;16(9):e1008229. doi: 10.1371/journal.pcbi.1008229. eCollection 2020 Sep. PLoS Comput Biol. 2020. PMID: 32936825 Free PMC article.
-
Structure-Based Approaches for Protein-Protein Interaction Prediction Using Machine Learning and Deep Learning.Biomolecules. 2025 Jan 17;15(1):141. doi: 10.3390/biom15010141. Biomolecules. 2025. PMID: 39858535 Free PMC article. Review.
-
CEGSO: Boosting Essential Proteins Prediction by Integrating Protein Complex, Gene Expression, Gene Ontology, Subcellular Localization and Orthology Information.Interdiscip Sci. 2021 Sep;13(3):349-361. doi: 10.1007/s12539-021-00426-7. Epub 2021 Mar 27. Interdiscip Sci. 2021. PMID: 33772722 Review.
Cited by
-
Artificial intelligence and machine learning applications for cultured meat.Front Artif Intell. 2024 Sep 24;7:1424012. doi: 10.3389/frai.2024.1424012. eCollection 2024. Front Artif Intell. 2024. PMID: 39381621 Free PMC article. Review.
-
AttentionEP: Predicting essential proteins via fusion of multiscale features by attention mechanisms.Comput Struct Biotechnol J. 2024 Nov 29;23:4315-4323. doi: 10.1016/j.csbj.2024.11.039. eCollection 2024 Dec. Comput Struct Biotechnol J. 2024. PMID: 39697678 Free PMC article.
-
A deep ensemble framework for human essential gene prediction by integrating multi-omics data.Sci Rep. 2025 Jul 21;15(1):26407. doi: 10.1038/s41598-025-99164-9. Sci Rep. 2025. PMID: 40691502 Free PMC article.
-
A seed expansion-based method to identify essential proteins by integrating protein-protein interaction sub-networks and multiple biological characteristics.BMC Bioinformatics. 2023 Nov 30;24(1):452. doi: 10.1186/s12859-023-05583-8. BMC Bioinformatics. 2023. PMID: 38036960 Free PMC article.
-
ECDEP: identifying essential proteins based on evolutionary community discovery and subcellular localization.BMC Genomics. 2024 Jan 26;25(1):117. doi: 10.1186/s12864-024-10019-5. BMC Genomics. 2024. PMID: 38279081 Free PMC article.
References
MeSH terms
Substances
Grants and funding
- 2021d06050003/The key scientific and technological breakthroughs in Anhui Province "Innovation of excellent wheat germplasm resources, discovery of important new genes and application in wheat molecular design breeding"
- 2021d06050003/The key scientific and technological breakthroughs in Anhui Province "Innovation of excellent wheat germplasm resources, discovery of important new genes and application in wheat molecular design breeding"
- 2021d06050003/The key scientific and technological breakthroughs in Anhui Province "Innovation of excellent wheat germplasm resources, discovery of important new genes and application in wheat molecular design breeding"
- 2021d06050003/The key scientific and technological breakthroughs in Anhui Province "Innovation of excellent wheat germplasm resources, discovery of important new genes and application in wheat molecular design breeding"
- 2021d06050003/The key scientific and technological breakthroughs in Anhui Province "Innovation of excellent wheat germplasm resources, discovery of important new genes and application in wheat molecular design breeding"
- 2021d06050003/The key scientific and technological breakthroughs in Anhui Province "Innovation of excellent wheat germplasm resources, discovery of important new genes and application in wheat molecular design breeding"
- 2021d06050003/The key scientific and technological breakthroughs in Anhui Province "Innovation of excellent wheat germplasm resources, discovery of important new genes and application in wheat molecular design breeding"
- 2021d06050003/The key scientific and technological breakthroughs in Anhui Province "Innovation of excellent wheat germplasm resources, discovery of important new genes and application in wheat molecular design breeding"
- [2020]555/"Three Renewal and One Creation"Innovation Platform Fund-Anhui Provincial Engineering Laboratory for Beidou Precision Agriculture lnformation (Anhui Developmentand Reform Innovation)
- [2020]555/"Three Renewal and One Creation"Innovation Platform Fund-Anhui Provincial Engineering Laboratory for Beidou Precision Agriculture lnformation (Anhui Developmentand Reform Innovation)
- [2020]555/"Three Renewal and One Creation"Innovation Platform Fund-Anhui Provincial Engineering Laboratory for Beidou Precision Agriculture lnformation (Anhui Developmentand Reform Innovation)
- [2020]555/"Three Renewal and One Creation"Innovation Platform Fund-Anhui Provincial Engineering Laboratory for Beidou Precision Agriculture lnformation (Anhui Developmentand Reform Innovation)
- [2020]555/"Three Renewal and One Creation"Innovation Platform Fund-Anhui Provincial Engineering Laboratory for Beidou Precision Agriculture lnformation (Anhui Developmentand Reform Innovation)
- [2020]555/"Three Renewal and One Creation"Innovation Platform Fund-Anhui Provincial Engineering Laboratory for Beidou Precision Agriculture lnformation (Anhui Developmentand Reform Innovation)
- [2020]555/"Three Renewal and One Creation"Innovation Platform Fund-Anhui Provincial Engineering Laboratory for Beidou Precision Agriculture lnformation (Anhui Developmentand Reform Innovation)
- [2020]555/"Three Renewal and One Creation"Innovation Platform Fund-Anhui Provincial Engineering Laboratory for Beidou Precision Agriculture lnformation (Anhui Developmentand Reform Innovation)
- SKLTOF20150103/The Open Fund of State Key Laboratory of Tea Plant Biology and Utilization
- SKLTOF20150103/The Open Fund of State Key Laboratory of Tea Plant Biology and Utilization
- SKLTOF20150103/The Open Fund of State Key Laboratory of Tea Plant Biology and Utilization
- SKLTOF20150103/The Open Fund of State Key Laboratory of Tea Plant Biology and Utilization
- SKLTOF20150103/The Open Fund of State Key Laboratory of Tea Plant Biology and Utilization
- SKLTOF20150103/The Open Fund of State Key Laboratory of Tea Plant Biology and Utilization
- SKLTOF20150103/The Open Fund of State Key Laboratory of Tea Plant Biology and Utilization
- SKLTOF20150103/The Open Fund of State Key Laboratory of Tea Plant Biology and Utilization
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

