Engineering drought and salinity tolerance traits in crops through CRISPR-mediated genome editing: Targets, tools, challenges, and perspectives
- PMID: 35927945
- PMCID: PMC9700172
- DOI: 10.1016/j.xplc.2022.100417
Engineering drought and salinity tolerance traits in crops through CRISPR-mediated genome editing: Targets, tools, challenges, and perspectives
Abstract
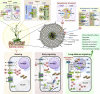
Prolonged periods of drought triggered by climate change hamper plant growth and cause substantial agricultural yield losses every year. In addition to drought, salinity is one of the major abiotic stresses that severely affect crop health and agricultural production. Plant responses to drought and salinity involve multiple processes that operate in a spatiotemporal manner, such as stress sensing, perception, epigenetic modifications, transcription, post-transcriptional processing, translation, and post-translational changes. Consequently, drought and salinity stress tolerance are polygenic traits influenced by genome-environment interactions. One of the ideal solutions to these challenges is the development of high-yielding crop varieties with enhanced stress tolerance, together with improved agricultural practices. Recently, genome-editing technologies, especially clustered regularly interspaced short palindromic repeats (CRISPR) tools, have been effectively applied to elucidate how plants deal with drought and saline environments. In this work, we aim to portray that the combined use of CRISPR-based genome engineering tools and modern genomic-assisted breeding approaches are gaining momentum in identifying genetic determinants of complex traits for crop improvement. This review provides a synopsis of plant responses to drought and salinity stresses at the morphological, physiological, and molecular levels. We also highlight recent advances in CRISPR-based tools and their use in understanding the multi-level nature of plant adaptations to drought and salinity stress. Integrating CRISPR tools with modern breeding approaches is ideal for identifying genetic factors that regulate plant stress-response pathways and for the introgression of beneficial traits to develop stress-resilient crops.
Keywords: CRISPR/Cas; drought tolerance; genome editing; polygenic traits; salt tolerance; trait introgression.
Copyright © 2022 The Author(s). Published by Elsevier Inc. All rights reserved.
Figures





Similar articles
-
Engineering abiotic stress tolerance via CRISPR/ Cas-mediated genome editing.J Exp Bot. 2020 Jan 7;71(2):470-479. doi: 10.1093/jxb/erz476. J Exp Bot. 2020. PMID: 31644801
-
State-of-the-Art in CRISPR Technology and Engineering Drought, Salinity, and Thermo-tolerant crop plants.Plant Cell Rep. 2022 Mar;41(3):815-831. doi: 10.1007/s00299-021-02681-w. Epub 2021 Mar 19. Plant Cell Rep. 2022. PMID: 33742256 Review.
-
CRISPR-Cas9-based genetic engineering for crop improvement under drought stress.Bioengineered. 2021 Dec;12(1):5814-5829. doi: 10.1080/21655979.2021.1969831. Bioengineered. 2021. PMID: 34506262 Free PMC article. Review.
-
Potential Application of CRISPR/Cas9 System to Engineer Abiotic Stress Tolerance in Plants.Protein Pept Lett. 2021;28(8):861-877. doi: 10.2174/0929866528666210218220138. Protein Pept Lett. 2021. PMID: 33602066 Review.
-
CRISPR/Cas9 mediated genome editing for crop improvement against Abiotic stresses: current trends and prospects.Funct Integr Genomics. 2024 Oct 25;24(6):199. doi: 10.1007/s10142-024-01480-2. Funct Integr Genomics. 2024. PMID: 39453513 Review.
Cited by
-
CRISPR/Cas Technology Revolutionizes Crop Breeding.Plants (Basel). 2023 Aug 30;12(17):3119. doi: 10.3390/plants12173119. Plants (Basel). 2023. PMID: 37687368 Free PMC article. Review.
-
Unravelling the molecular mechanism underlying drought stress tolerance in Dinanath (Pennisetum pedicellatum Trin.) grass via integrated transcriptomic and metabolomic analyses.BMC Plant Biol. 2024 Oct 5;24(1):928. doi: 10.1186/s12870-024-05579-3. BMC Plant Biol. 2024. PMID: 39367330 Free PMC article.
-
Bacillus subtilis ER-08, a multifunctional plant growth-promoting rhizobacterium, promotes the growth of fenugreek (Trigonella foenum-graecum L.) plants under salt and drought stress.Front Microbiol. 2023 Aug 24;14:1208743. doi: 10.3389/fmicb.2023.1208743. eCollection 2023. Front Microbiol. 2023. PMID: 37692403 Free PMC article.
-
Low-cost and reliable substrate-based phenotyping platform for screening salt tolerance of cutting propagation-dependent grass, paspalum vaginatum.Plant Methods. 2024 Jun 19;20(1):94. doi: 10.1186/s13007-024-01225-z. Plant Methods. 2024. PMID: 38898477 Free PMC article.
-
Research Advancements in Salt Tolerance of Cucurbitaceae: From Salt Response to Molecular Mechanisms.Int J Mol Sci. 2024 Aug 21;25(16):9051. doi: 10.3390/ijms25169051. Int J Mol Sci. 2024. PMID: 39201741 Free PMC article. Review.
References
-
- Agarwal G., Kudapa H., Ramalingam A., Choudhary D., Sinha P., Garg V., Singh V.K., Patil G.B., Pandey M.K., Nguyen H.T., et al. Epigenetics and epigenomics: underlying mechanisms, relevance, and implications in crop improvement. Funct. Integr. Genomics. 2020;20:739–761. - PubMed
-
- Ahmad S., Tang L., Shahzad R., Mawia A.M., Rao G.S., Jamil S., Wei C., Sheng Z., Shao G., Wei X., et al. CRISPR-based crop improvements: a way forward to achieve zero hunger. J. Agric. Food Chem. 2021;69:8307–8323. - PubMed
-
- Alfatih A., Wu J., Jan S.U., Zhang Z.S., Xia J.Q., Xiang C.B. Loss of rice PARAQUAT TOLERANCE 3 confers enhanced resistance to abiotic stresses and increases grain yield in field. Plant Cell Environ. 2020;43:2743–2754. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

