A modified CUT&RUN-seq technique for qPCR analysis of chromatin-protein interactions
- PMID: 35928003
- PMCID: PMC9344018
- DOI: 10.1016/j.xpro.2022.101529
A modified CUT&RUN-seq technique for qPCR analysis of chromatin-protein interactions
Abstract
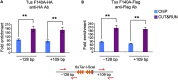
Chromatin immunoprecipitation coupled with quantitative PCR (ChIP-qPCR) even with optimization may give low signal-to-background ratio and spatial resolution. Here, we adapted Cleavage Under Targets and Release Using Nuclease (CUT&RUN) (originally developed by the Henikoff group) to develop CUT&RUN-qPCR. By studying the recruitment of selected proteins (but amenable to other proteins), we find that CUT&RUN-qPCR is more sensitive and gives better spatial resolution than ChIP-qPCR. For complete details on the use and execution of this protocol, please refer to Skene et al. (2018) and Skene and Henikoff (2017).
Keywords: Cell Biology; Cell-based Assays; Chromatin immunoprecipitation (ChIP); Molecular Biology.
© 2022 The Authors.
Conflict of interest statement
The authors declare no competing interests.
Figures






References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

