Protocols for protein-DNA binding analysis of a zinc finger transcription factor bound to its cognate promoter
- PMID: 35928006
- PMCID: PMC9344028
- DOI: 10.1016/j.xpro.2022.101598
Protocols for protein-DNA binding analysis of a zinc finger transcription factor bound to its cognate promoter
Abstract
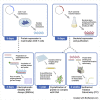
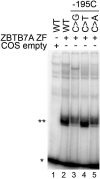
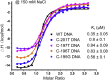
Here, we describe protocols to interrogate the binding of the zinc fingers of the transcription factor ZBTB7A to the fetal γ-globin (HBG) promoter. We detail the steps for performing electrophoretic mobility shift assays (EMSAs), X-ray crystallography, and isothermal titration calorimetry (ITC) to explore this interaction. These techniques could readily be applied to the structural studies of other zinc finger transcription factors and cognate DNA sequences. For complete details on the use and execution of this protocol, please refer to Yang et al. (2021).
Keywords: Biophysics; Molecular Biology; Protein Biochemistry; Protein expression and purification; X-ray Crystallography.
© 2022 The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



References
-
- Amano R., Furukawa T., Sakamoto T. ITC measurement for high-affinity aptamers binding to their target proteins. Methods Mol. Biol. 2019;1964:119–128. - PubMed
-
- Collaborative Computational Project N. The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr. 1994;50:760–763. - PubMed
-
- Emsley P., Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 2004;60:2126–2132. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

