Phylogenetic analysis and accessory genome diversity reveal insight into the evolutionary history of Streptococcus dysgalactiae
- PMID: 35928143
- PMCID: PMC9343751
- DOI: 10.3389/fmicb.2022.952110
Phylogenetic analysis and accessory genome diversity reveal insight into the evolutionary history of Streptococcus dysgalactiae
Abstract
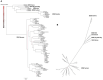
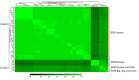
Streptococcus dysgalactiae (SD) is capable of infecting both humans and animals and causing a wide range of invasive and non-invasive infections. With two subspecies, the taxonomic status of subspecies of SD remains controversial. Subspecies equisimilis (SDSE) is an important human pathogen, while subspecies dysgalactiae (SDSD) has been considered a strictly animal pathogen; however, occasional human infections by this subspecies have been reported in the last few years. Moreover, the differences between the adaptation of SDSD within humans and other animals are still unknown. In this work, we provide a phylogenomic analysis based on the single-copy core genome of 106 isolates from both the subspecies and different infected hosts (animal and human hosts). The accessory genome of this species was also analyzed for screening of genes that could be specifically involved with adaptation to different hosts. Additionally, we searched putatively adaptive traits among prophage regions to infer the importance of transduction in the adaptation of SD to different hosts. Core genome phylogenetic relationships segregate all human SDSE in a single cluster separated from animal SD isolates. The subgroup of bovine SDSD evolved from this later clade and harbors a specialized accessory genome characterized by the presence of specific virulence determinants (e.g., cspZ) and carbohydrate metabolic functions (e.g., fructose operon). Together, our results indicate a host-specific SD and the existence of an SDSD group that causes human-animal cluster infections may be due to opportunistic infections, and that the exact incidence of SDSD human infections may be underestimated due to failures in identification based on the hemolytic patterns. However, more detailed research into the isolation of human SD is needed to assess whether it is a carrier phenomenon or whether the species can be permanently integrated into the human microbiome, making it ready to cause opportunistic infections.
Keywords: SDSD and SDSE; Streptococcus dysgalactiae; accessory genome; core-genome; prophages regions.
Copyright © 2022 Alves-Barroco, Brito, Santos-Sanches and Fernandes.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures




Similar articles
-
New Insights on Streptococcus dysgalactiae subsp. dysgalactiae Isolates.Front Microbiol. 2021 Jul 15;12:686413. doi: 10.3389/fmicb.2021.686413. eCollection 2021. Front Microbiol. 2021. PMID: 34335512 Free PMC article.
-
Whole genome sequencing reveals possible host species adaptation of Streptococcus dysgalactiae.Sci Rep. 2021 Aug 30;11(1):17350. doi: 10.1038/s41598-021-96710-z. Sci Rep. 2021. PMID: 34462475 Free PMC article.
-
Comparative genomic analysis of Streptococcus dysgalactiae subspecies dysgalactiae, an occasional cause of zoonotic infection.Pathology. 2020 Feb;52(2):262-266. doi: 10.1016/j.pathol.2019.09.016. Epub 2019 Dec 16. Pathology. 2020. PMID: 31859005
-
Intra-familial transmission of Streptococcus dysgalactiae subsp. equisimilis (SDSE): A first case report and review of the literature.J Infect Chemother. 2022 Jun;28(6):819-822. doi: 10.1016/j.jiac.2022.01.017. Epub 2022 Feb 1. J Infect Chemother. 2022. PMID: 35115237 Review.
-
Severe invasive streptococcal infection by Streptococcus pyogenes and Streptococcus dysgalactiae subsp. equisimilis.Microbiol Immunol. 2016 Jan;60(1):1-9. doi: 10.1111/1348-0421.12334. Microbiol Immunol. 2016. PMID: 26762200 Review.
Cited by
-
The Genetic Diversity and Antimicrobial Resistance of Pyogenic Pathogens Isolated from Porcine Lymph Nodes.Antibiotics (Basel). 2023 Jun 7;12(6):1026. doi: 10.3390/antibiotics12061026. Antibiotics (Basel). 2023. PMID: 37370345 Free PMC article.
-
Genetic analysis reveals the genetic diversity and zoonotic potential of Streptococcus dysgalactiae isolates from sheep.Sci Rep. 2025 Jan 25;15(1):3165. doi: 10.1038/s41598-025-87781-3. Sci Rep. 2025. PMID: 39863800 Free PMC article.
-
Integrative genomic, virulence, and transcriptomic analysis of emergent Streptococcus dysgalactiae subspecies equisimilis (SDSE) emm type stG62647 isolates causing human infections.mBio. 2024 Nov 13;15(11):e0257824. doi: 10.1128/mbio.02578-24. Epub 2024 Oct 17. mBio. 2024. PMID: 39417630 Free PMC article.
-
Evolutionary dynamics of the accessory genomes of Staphylococcus aureus.mSphere. 2024 Apr 23;9(4):e0075123. doi: 10.1128/msphere.00751-23. Epub 2024 Mar 19. mSphere. 2024. PMID: 38501935 Free PMC article.
-
Streptococcus dysgalactiae subsp. equisimilis infection and its intersection with Streptococcus pyogenes.Clin Microbiol Rev. 2024 Sep 12;37(3):e0017523. doi: 10.1128/cmr.00175-23. Epub 2024 Jun 10. Clin Microbiol Rev. 2024. PMID: 38856686 Free PMC article. Review.
References
-
- Abdelsalam M., Asheg A., Eissa A. E. (2013). Streptococcus dysgalactiae: an emerging pathogen of fishes and mammals. Int. J. Vet. Sci. Med. 1 1–6. 10.1016/j.ijvsm.2013.04.002 - DOI
-
- Alves-Barroco C., Roma-Rodrigues C., Raposo L. R., Brás C., Diniz M., Caço J., et al. (2018). Streptococcus dysgalactiae subsp. dysgalactiae isolated from milk of the bovine udder as emerging pathogens: in vitro and in vivo infection of human cells and zebrafish as biological models. MicrobiologyOpen 8:e00623. 10.1002/mbo3.623 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

