YWHAG inhibits influenza a virus replication by suppressing the release of viral M2 protein
- PMID: 35928168
- PMCID: PMC9343881
- DOI: 10.3389/fmicb.2022.951009
YWHAG inhibits influenza a virus replication by suppressing the release of viral M2 protein
Abstract
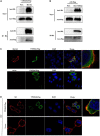
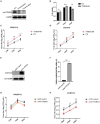
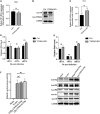
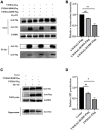
Influenza A virus (IAV) poses a serious threat to human life and property. The IAV matrix protein 2 (M2) is significant in viral budding. Increasing studies have proven the important roles of host factors in IAV replication. In this study, immunoprecipitation combined with mass spectrometry revealed that the host protein tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein gamma (YWHAG), which belongs to the 14-3-3 protein scaffold family, interacts with M2. Their interactions were further confirmed by co-immunoprecipitation (Co-IP), immunofluorescence, and confocal microscopy of virus-infected HeLa cells. Moreover, we constructed YWHAG-KO and YWHAG-overexpressing cells and found that YWHAG knockout significantly increased viral production, whereas its overexpression reduced the titer of virus progeny. Therefore, YWHAG is a negative regulatory factor during IAV infection. Further, YWHAG knockout or overexpression had no effect on the binding, entry, or viral RNA replication in the early stages of the virus life cycle. On the contrary, it impaired the release of virions at the plasma membrane as determined using transmission electron microscopy and suppressed the M2-mediated budding of the influenza virus. Importantly, the H158F mutation of YWHAG was found to affect interaction with M2 and its budding. Collectively, our work demonstrates that YWHAG is a novel cellular regulator that targets and mediates the interaction and release of M2.
Keywords: M2 protein; YWHAG; influenza A virus; protein–protein interaction; viral budding.
Copyright © 2022 Mao, Cao, Xu, Xia, Ren, Han, Li, Hui, Lin, Huang and Jin.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

Similar articles
-
PSMD12-Mediated M1 Ubiquitination of Influenza A Virus at K102 Regulates Viral Replication.J Virol. 2022 Aug 10;96(15):e0078622. doi: 10.1128/jvi.00786-22. Epub 2022 Jul 21. J Virol. 2022. PMID: 35861516 Free PMC article.
-
Human annexin A6 interacts with influenza a virus protein M2 and negatively modulates infection.J Virol. 2012 Feb;86(3):1789-801. doi: 10.1128/JVI.06003-11. Epub 2011 Nov 23. J Virol. 2012. PMID: 22114333 Free PMC article.
-
Autophagy Promotes Replication of Influenza A Virus In Vitro.J Virol. 2019 Feb 5;93(4):e01984-18. doi: 10.1128/JVI.01984-18. Print 2019 Feb 15. J Virol. 2019. PMID: 30541828 Free PMC article.
-
A Defect in Influenza A Virus Particle Assembly Specific to Primary Human Macrophages.mBio. 2018 Oct 23;9(5):e01916-18. doi: 10.1128/mBio.01916-18. mBio. 2018. PMID: 30352935 Free PMC article.
-
An overview of influenza A virus genes, protein functions, and replication cycle highlighting important updates.Virus Genes. 2022 Aug;58(4):255-269. doi: 10.1007/s11262-022-01904-w. Epub 2022 Apr 26. Virus Genes. 2022. PMID: 35471490 Review.
Cited by
-
Cholesterol and M2 Rendezvous in Budding and Scission of Influenza A Virus.Subcell Biochem. 2023;106:441-459. doi: 10.1007/978-3-031-40086-5_16. Subcell Biochem. 2023. PMID: 38159237
-
Integrating bulk and single-cell data to predict the prognosis and identify the immune landscape in HNSCC.J Cell Mol Med. 2024 Jan;28(1):e18009. doi: 10.1111/jcmm.18009. Epub 2023 Oct 26. J Cell Mol Med. 2024. PMID: 37882107 Free PMC article.
References
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

