Activation of STING Based on Its Structural Features
- PMID: 35928815
- PMCID: PMC9343627
- DOI: 10.3389/fimmu.2022.808607
Activation of STING Based on Its Structural Features
Abstract
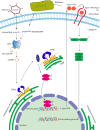
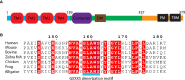
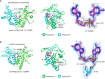
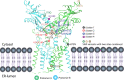
The cGAS-cGAMP-STING pathway is an important innate immune signaling cascade responsible for the sensing of abnormal cytosolic double-stranded DNA (dsDNA), which is a hallmark of infection or cancers. Recently, tremendous progress has been made in the understanding of the STING activation mechanism from various aspects. In this review, the molecular mechanism of activation of STING protein based on its structural features is briefly discussed. The underlying molecular mechanism of STING activation will enable us to develop novel therapeutics to treat STING-associated diseases and understand how STING has evolved to eliminate infection and maintain immune homeostasis in innate immunity.
Keywords: cyclic GMP-AMP; cyclic GMP-AMP Synthase (cGAS); cyclic dinucleotide; innate immunity; stimulator of interferon genes.
Copyright © 2022 Hussain, Xie, Jabeen, Lu, Yang, Wu and Shang.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

