Interplay Between Immune and Cancer-Associated Fibroblasts: A Path to Target Metalloproteinases in Penile Cancer
- PMID: 35928876
- PMCID: PMC9343588
- DOI: 10.3389/fonc.2022.935093
Interplay Between Immune and Cancer-Associated Fibroblasts: A Path to Target Metalloproteinases in Penile Cancer
Abstract
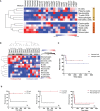
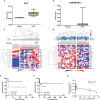
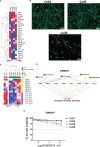
Extracellular matrix (ECM) remodeling and inflammation have been reported in penile carcinomas (PeCa). However, the cell types and cellular crosstalk involved in PeCa are unexplored. We aimed to characterize the complexity of cells and pathways involved in the tumor microenvironment (TME) in PeCa and propose target molecules associated with the TME. We first investigated the prognostic impact of cell types with a secretory profile to identify drug targets that modulate TME-enriched cells. The secretome analysis using the PeCa transcriptome revealed the enrichment of inflammation and extracellular matrix pathways. Twenty-three secreted factors were upregulated, mainly collagens and matrix metalloproteinases (MMPs). The deregulation of collagens and MMPs was confirmed by Quantitative reverse transcription - polymerase chain reaction (RT-qPCR). Further, the deconvolution method (digital cytometry) of the bulk samples revealed a high proportion of macrophages and dendritic cells (DCs) and B cells. Increased DCs and B cells were associated with better survival. A high proportion of cancer-associated fibroblasts (CAFs) was observed in low-survival patients. Patients with increased CAFs had decreased immune cell proportions. The treatment with the MMP inhibitor GM6001 in CAF cells derived from PeCa resulted in altered cell viability. We reported a crosstalk between immune cells and CAFs, and the proportion of these cell populations was associated with prognosis. We demonstrate that a drug targeting MMPs modulates CAFs, expanding the therapeutic options of PeCa.
Keywords: cancer-associated fibroblasts; penile cancer; response to therapy; secretome; transcriptome.
Copyright © 2022 Cury, Kuasne, Souza, Muñoz, da Silva, Lopes, Scapulatempo-Neto, Faria, Delaissé, Marchi and Rogatto.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


References
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

