Discovery of nonautonomous modulators of activated Ras
- PMID: 35929788
- PMCID: PMC9526067
- DOI: 10.1093/g3journal/jkac200
Discovery of nonautonomous modulators of activated Ras
Abstract
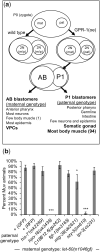
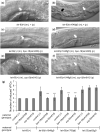
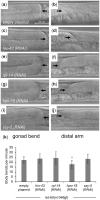
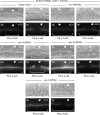
Communication between mesodermal cells and epithelial cells is fundamental to normal animal development and is frequently disrupted in cancer. However, the genes and processes that mediate this communication are incompletely understood. To identify genes that mediate this communication and alter the proliferation of cells with an oncogenic Ras genotype, we carried out a tissue-specific genome-wide RNAi screen in Caenorhabditis elegans animals bearing a let-60(n1046gf) (RasG13E) allele. The screen identifies 24 genes that, when knocked down in adjacent mesodermal tissue, suppress the increased vulval epithelial cell proliferation defect associated with let-60(n1046gf). Importantly, gene knockdown reverts the mutant animals to a wild-type phenotype. Using chimeric animals, we genetically confirm that 2 of the genes function nonautonomously to revert the let-60(n1046gf) phenotype. The effect is genotype restricted, as knockdown does not alter development in a wild type (let-60(+)) or activated EGF receptor (let-23(sa62gf)) background. Although many of the genes identified encode proteins involved in essential cellular processes, including chromatin formation, ribosome function, and mitochondrial ATP metabolism, knockdown does not alter the normal development or function of targeted mesodermal tissues, indicating that the phenotype derives from specific functions performed by these cells. We show that the genes act in a manner distinct from 2 signal ligand classes (EGF and Wnt) known to influence the development of vulval epithelial cells. Altogether, the results identify genes with a novel function in mesodermal cells required for communicating with and promoting the proliferation of adjacent epithelial cells with an activated Ras genotype.
Keywords: Caenorhabditis elegans; ATP synthase; Ras; cell communication; vulval development.
© The Author(s) 2022. Published by Oxford University Press on behalf of Genetics Society of America.
Figures


Similar articles
-
Reciprocal EGF signaling back to the uterus from the induced C. elegans vulva coordinates morphogenesis of epithelia.Curr Biol. 1999 Mar 11;9(5):237-46. doi: 10.1016/s0960-9822(99)80112-2. Curr Biol. 1999. PMID: 10074449
-
Inhibition of Caenorhabditis elegans vulval induction by gap-1 and by let-23 receptor tyrosine kinase.Genes Dev. 1997 Oct 15;11(20):2715-28. doi: 10.1101/gad.11.20.2715. Genes Dev. 1997. PMID: 9334333 Free PMC article.
-
The Caenorhabditis elegans synthetic multivulva genes prevent ras pathway activation by tightly repressing global ectopic expression of lin-3 EGF.PLoS Genet. 2011 Dec;7(12):e1002418. doi: 10.1371/journal.pgen.1002418. Epub 2011 Dec 29. PLoS Genet. 2011. PMID: 22242000 Free PMC article.
-
LET-23-mediated signal transduction during Caenorhabditis elegans development.Mol Reprod Dev. 1995 Dec;42(4):523-8. doi: 10.1002/mrd.1080420422. Mol Reprod Dev. 1995. PMID: 8607985 Review.
-
Signal transduction and cell fate specification during Caenorhabditis elegans vulval development.Curr Opin Genet Dev. 1994 Aug;4(4):508-16. doi: 10.1016/0959-437x(94)90065-b. Curr Opin Genet Dev. 1994. PMID: 7950317 Review.
Cited by
-
Caenorhabditis elegans for research on cancer hallmarks.Dis Model Mech. 2023 Jun 1;16(6):dmm050079. doi: 10.1242/dmm.050079. Epub 2023 Jun 6. Dis Model Mech. 2023. PMID: 37278614 Free PMC article.
-
Functional distinction in oncogenic Ras variant activity in Caenorhabditis elegans.Dis Model Mech. 2024 Aug 1;17(8):dmm050577. doi: 10.1242/dmm.050577. Epub 2024 Aug 14. Dis Model Mech. 2024. PMID: 38946472 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
