Preclinical Characterization of AZD5305, A Next-Generation, Highly Selective PARP1 Inhibitor and Trapper
- PMID: 35929986
- PMCID: PMC9623235
- DOI: 10.1158/1078-0432.CCR-22-0301
Preclinical Characterization of AZD5305, A Next-Generation, Highly Selective PARP1 Inhibitor and Trapper
Abstract
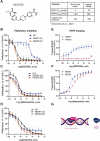
Purpose: We hypothesized that inhibition and trapping of PARP1 alone would be sufficient to achieve antitumor activity. In particular, we aimed to achieve selectivity over PARP2, which has been shown to play a role in the survival of hematopoietic/stem progenitor cells in animal models. We developed AZD5305 with the aim of achieving improved clinical efficacy and wider therapeutic window. This next-generation PARP inhibitor (PARPi) could provide a paradigm shift in clinical outcomes achieved by first-generation PARPi, particularly in combination.
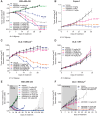
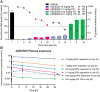
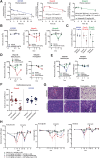
Experimental design: AZD5305 was tested in vitro for PARylation inhibition, PARP-DNA trapping, and antiproliferative abilities. In vivo efficacy was determined in mouse xenograft and PDX models. The potential for hematologic toxicity was evaluated in rat models, as monotherapy and combination.
Results: AZD5305 is a highly potent and selective inhibitor of PARP1 with 500-fold selectivity for PARP1 over PARP2. AZD5305 inhibits growth in cells with deficiencies in DNA repair, with minimal/no effects in other cells. Unlike first-generation PARPi, AZD5305 has minimal effects on hematologic parameters in a rat pre-clinical model at predicted clinically efficacious exposures. Animal models treated with AZD5305 at doses ≥0.1 mg/kg once daily achieved greater depth of tumor regression compared to olaparib 100 mg/kg once daily, and longer duration of response.
Conclusions: AZD5305 potently and selectively inhibits PARP1 resulting in excellent antiproliferative activity and unprecedented selectivity for DNA repair deficient versus proficient cells. These data confirm the hypothesis that targeting only PARP1 can retain the therapeutic benefit of nonselective PARPi, while reducing potential for hematotoxicity. AZD5305 is currently in phase I trials (NCT04644068).
©2022 The Authors; Published by the American Association for Cancer Research.
Figures


References
-
- Johannes JW, Balazs A, Barratt D, Bista M, Chuba MD, Cosulich S, et al. Discovery of 5-{4-[(7-Ethyl-6-oxo-5,6-dihydro-1,5-naphthyridin-3-yl)methyl]piperazin-1-yl}-N-m ethylpyridine-2-carboxamide (AZD5305): a PARP1-DNA trapper with high selectivity for PARP1 over PARP2 and other PARPs. J Med Chem 2021;64:14498–512. - PubMed
Publication types
MeSH terms
Substances
Associated data
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

