Genomic dissection of the Vibrio cholerae O-serogroup global reference strains: reassessing our view of diversity and plasticity between two chromosomes
- PMID: 35930328
- PMCID: PMC9484750
- DOI: 10.1099/mgen.0.000860
Genomic dissection of the Vibrio cholerae O-serogroup global reference strains: reassessing our view of diversity and plasticity between two chromosomes
Abstract
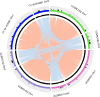
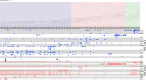
Approximately 200 O-serogroups of Vibrio cholerae have already been identified; however, only 2 serogroups, O1 and O139, are strongly related to pandemic cholera. The study of non-O1 and non-O139 strains has hitherto been limited. Nevertheless, there are other clinically and epidemiologically important serogroups causing outbreaks with cholera-like disease. Here, we report a comprehensive genome analysis of the whole set of V. cholerae O-serogroup reference strains to provide an overview of this important bacterial pathogen. It revealed structural diversity of the O-antigen biosynthesis gene clusters located at specific loci on chromosome 1 and 16 pairs of strains with almost identical O-antigen biosynthetic gene clusters but differing in serological patterns. This might be due to the presence of O-antigen biosynthesis-related genes at secondary loci on chromosome 2.
Keywords: O-antigen biosynthetic gene cluster; O-serogroup reference strain; Vibrio cholerae; multi-chromosomal bacteria.
Conflict of interest statement
The authors declare that there are no conflicts of interest.
Figures



References
-
- World Health Orginization Outbreak of gastro-enteritis by non agglutinable (NAG) vibrios = épidémie de gastro-entérite due a des vibrions non agglutinables. Weekly Epidemiological Record. 1969;44:10.

