The TPR domain of PgaA is a multifunctional scaffold that binds PNAG and modulates PgaB-dependent polymer processing
- PMID: 35930610
- PMCID: PMC9384988
- DOI: 10.1371/journal.ppat.1010750
The TPR domain of PgaA is a multifunctional scaffold that binds PNAG and modulates PgaB-dependent polymer processing
Abstract
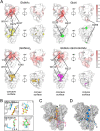
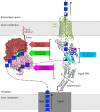
The synthesis of exopolysaccharides as biofilm matrix components by pathogens is a crucial factor for chronic infections and antibiotic resistance. Many periplasmic proteins involved in polymer processing and secretion in Gram-negative synthase dependent exopolysaccharide biosynthetic systems have been individually characterized. The operons responsible for the production of PNAG, alginate, cellulose and the Pel polysaccharide each contain a gene that encodes an outer membrane associated tetratricopeptide repeat (TPR) domain containing protein. While the TPR domain has been shown to bind other periplasmic proteins, the functional consequences of these interactions for the polymer remain poorly understood. Herein, we show that the C-terminal TPR region of PgaA interacts with the de-N-acetylase domain of PgaB, and increases its deacetylase activity. Additionally, we found that when the two proteins form a complex, the glycoside hydrolase activity of PgaB is also increased. To better understand structure-function relationships we determined the crystal structure of a stable TPR module, which has a conserved groove formed by three repeat motifs. Tryptophan quenching, mass spectrometry analysis and molecular dynamics simulation studies suggest that the crystallized TPR module can bind PNAG/dPNAG via its electronegative groove on the concave surface, and potentially guide the polymer through the periplasm towards the porin for export. Our results suggest a scaffolding role for the TPR domain that combines PNAG/dPNAG translocation with the modulation of its chemical structure by PgaB.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures





Similar articles
-
Structural Basis for Translocation of a Biofilm-supporting Exopolysaccharide across the Bacterial Outer Membrane.J Biol Chem. 2016 May 6;291(19):10046-57. doi: 10.1074/jbc.M115.711762. Epub 2016 Mar 8. J Biol Chem. 2016. PMID: 26957546 Free PMC article.
-
Modification and periplasmic translocation of the biofilm exopolysaccharide poly-β-1,6-N-acetyl-D-glucosamine.Proc Natl Acad Sci U S A. 2014 Jul 29;111(30):11013-8. doi: 10.1073/pnas.1406388111. Epub 2014 Jul 3. Proc Natl Acad Sci U S A. 2014. PMID: 24994902 Free PMC article.
-
PgaB orthologues contain a glycoside hydrolase domain that cleaves deacetylated poly-β(1,6)-N-acetylglucosamine and can disrupt bacterial biofilms.PLoS Pathog. 2018 Apr 23;14(4):e1006998. doi: 10.1371/journal.ppat.1006998. eCollection 2018 Apr. PLoS Pathog. 2018. PMID: 29684093 Free PMC article.
-
Synthase-dependent exopolysaccharide secretion in Gram-negative bacteria.Trends Microbiol. 2013 Feb;21(2):63-72. doi: 10.1016/j.tim.2012.10.001. Epub 2012 Oct 29. Trends Microbiol. 2013. PMID: 23117123 Free PMC article. Review.
-
Gram-negative synthase-dependent exopolysaccharide biosynthetic machines.Curr Opin Struct Biol. 2018 Dec;53:32-44. doi: 10.1016/j.sbi.2018.05.001. Epub 2018 May 26. Curr Opin Struct Biol. 2018. PMID: 29843050 Review.
Cited by
-
Molecular insights into phosphoethanolamine cellulose formation and secretion.bioRxiv [Preprint]. 2024 Apr 8:2024.04.04.588173. doi: 10.1101/2024.04.04.588173. bioRxiv. 2024. Update in: Nat Commun. 2024 Sep 6;15(1):7798. doi: 10.1038/s41467-024-51838-0. PMID: 38645035 Free PMC article. Updated. Preprint.
-
Spatio-genetically coordinated TPR domain-containing proteins modulate c-di-GMP signaling in Vibrio vulnificus.PLoS Pathog. 2025 Jul 16;21(7):e1013353. doi: 10.1371/journal.ppat.1013353. eCollection 2025 Jul. PLoS Pathog. 2025. PMID: 40668869 Free PMC article.
-
The biofilm community resurfaces: new findings and post-pandemic progress.J Bacteriol. 2023 Oct 26;205(10):e0016623. doi: 10.1128/jb.00166-23. Epub 2023 Sep 27. J Bacteriol. 2023. PMID: 37756166 Free PMC article. Review.
-
Transition transferases prime bacterial capsule polymerization.Nat Chem Biol. 2025 Jan;21(1):120-130. doi: 10.1038/s41589-024-01664-8. Epub 2024 Jul 1. Nat Chem Biol. 2025. PMID: 38951648 Free PMC article.
-
Structural and functional analysis of Pseudomonas aeruginosa PelA provides insight into the modification of the Pel exopolysaccharide.J Biol Chem. 2025 May;301(5):108432. doi: 10.1016/j.jbc.2025.108432. Epub 2025 Mar 20. J Biol Chem. 2025. PMID: 40120681 Free PMC article.
References
-
- Marmont LS, Whitfield GB, Rich JD, Yip P, Giesbrecht LB, Stremick CA, et al.. PelA and PelB form a modification and secretion complex essential for Pel polysaccharide-dependent biofilm formation in Pseudomonas aeruginosa. J Biological Chem. 2017;292: 19411–19422. doi: 10.1074/jbc.M117.812842 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

