Spatial resolution of an integrated C4+CAM photosynthetic metabolism
- PMID: 35930634
- PMCID: PMC9355352
- DOI: 10.1126/sciadv.abn2349
Spatial resolution of an integrated C4+CAM photosynthetic metabolism
Abstract
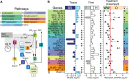
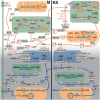
C4 and CAM photosynthesis have repeatedly evolved in plants over the past 30 million years. Because both repurpose the same set of enzymes but differ in their spatial and temporal deployment, they have long been considered as distinct and incompatible adaptations. Portulaca contains multiple C4 species that perform CAM when droughted. Spatially explicit analyses of gene expression reveal that C4 and CAM systems are completely integrated in Portulaca oleracea, with CAM and C4 carbon fixation occurring in the same cells and CAM-generated metabolites likely incorporated directly into the C4 cycle. Flux balance analysis corroborates the gene expression findings and predicts an integrated C4+CAM system under drought. This first spatially explicit description of a C4+CAM photosynthetic metabolism presents a potential new blueprint for crop improvement.
Figures





Similar articles
-
Gene co-expression reveals the modularity and integration of C4 and CAM in Portulaca.Plant Physiol. 2022 Jun 1;189(2):735-753. doi: 10.1093/plphys/kiac116. Plant Physiol. 2022. PMID: 35285495 Free PMC article.
-
Exploring C4-CAM plasticity within the Portulaca oleracea complex.Sci Rep. 2020 Aug 28;10(1):14237. doi: 10.1038/s41598-020-71012-y. Sci Rep. 2020. PMID: 32859905 Free PMC article.
-
A matter of time: regulatory events behind the synchronization of C4 and crassulacean acid metabolism in Portulaca oleracea.J Exp Bot. 2022 Aug 11;73(14):4867-4885. doi: 10.1093/jxb/erac163. J Exp Bot. 2022. PMID: 35439821
-
Achievable productivities of certain CAM plants: basis for high values compared with C3 and C4 plants.New Phytol. 1991 Oct;119(2):183-205. doi: 10.1111/j.1469-8137.1991.tb01022.x. New Phytol. 1991. PMID: 33874131 Review.
-
Facultative crassulacean acid metabolism (CAM) plants: powerful tools for unravelling the functional elements of CAM photosynthesis.J Exp Bot. 2014 Jul;65(13):3425-41. doi: 10.1093/jxb/eru063. Epub 2014 Mar 18. J Exp Bot. 2014. PMID: 24642847 Review.
Cited by
-
Molecular Mechanisms Underlying the Establishment and Maintenance of Vascular Stem Cells in Arabidopsis thaliana.Plant Cell Physiol. 2023 Mar 15;64(3):274-283. doi: 10.1093/pcp/pcac161. Plant Cell Physiol. 2023. PMID: 36398989 Free PMC article. Review.
-
Spatially resolved transcriptomic analysis of the germinating barley grain.Nucleic Acids Res. 2023 Aug 25;51(15):7798-7819. doi: 10.1093/nar/gkad521. Nucleic Acids Res. 2023. PMID: 37351575 Free PMC article.
-
The transcriptional integration of environmental cues with root cell type development.Plant Physiol. 2024 Dec 2;196(4):2150-2161. doi: 10.1093/plphys/kiae425. Plant Physiol. 2024. PMID: 39288006 Free PMC article. Review.
-
Synthetic crassulacean acid metabolism (SynCAM) for improving water-use efficiency in plants.Philos Trans R Soc Lond B Biol Sci. 2025 May 29;380(1927):20240249. doi: 10.1098/rstb.2024.0249. Epub 2025 May 29. Philos Trans R Soc Lond B Biol Sci. 2025. PMID: 40439297 Review.
-
The CAM lineages of planet Earth.Ann Bot. 2023 Nov 25;132(4):627-654. doi: 10.1093/aob/mcad135. Ann Bot. 2023. PMID: 37698538 Free PMC article.
References
-
- Edwards E. J., Evolutionary trajectories, accessibility and other metaphors: The case of C4 and CAM photosynthesis. New Phytol. 223, 1742–1755 (2019). - PubMed
-
- Sage R. F., Sage T. L., Kocacinar F., Photorespiration and the evolution of C4 photosynthesis. Rev. Plant Biol. 63, 19–47 (2012). - PubMed
-
- Black C. C., Chen T. M., Brown R. H., Biochemical basis for plant competition. Weed Sci. 17, 338–344 (1969).
-
- Osmond C. B., Crassulacean acid metabolism: A curiosity in context. Annu. Rev. Plant Physiol. 29, 379–414 (1978).
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

