Postmitotic accumulation of histone variant H3.3 in new cortical neurons establishes neuronal chromatin, transcriptome, and identity
- PMID: 35930666
- PMCID: PMC9371731
- DOI: 10.1073/pnas.2116956119
Postmitotic accumulation of histone variant H3.3 in new cortical neurons establishes neuronal chromatin, transcriptome, and identity
Abstract
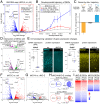
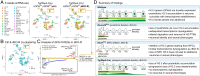
Histone variants, which can be expressed outside of S-phase and deposited DNA synthesis-independently, provide long-term histone replacement in postmitotic cells, including neurons. Beyond replenishment, histone variants also play active roles in gene regulation by modulating chromatin states or enabling nucleosome turnover. Here, we uncover crucial roles for the histone H3 variant H3.3 in neuronal development. We find that newborn cortical excitatory neurons, which have only just completed replication-coupled deposition of canonical H3.1 and H3.2, substantially accumulate H3.3 immediately postmitosis. Codeletion of H3.3-encoding genes H3f3a and H3f3b from newly postmitotic neurons abrogates H3.3 accumulation, markedly alters the histone posttranslational modification landscape, and causes widespread disruptions to the establishment of the neuronal transcriptome. These changes coincide with developmental phenotypes in neuronal identities and axon projections. Thus, preexisting, replication-dependent histones are insufficient for establishing neuronal chromatin and transcriptome; de novo H3.3 is required. Stage-dependent deletion of H3f3a and H3f3b from 1) cycling neural progenitor cells, 2) neurons immediately postmitosis, or 3) several days later, reveals the first postmitotic days to be a critical window for de novo H3.3. After H3.3 accumulation within this developmental window, codeletion of H3f3a and H3f3b does not lead to immediate H3.3 loss, but causes progressive H3.3 depletion over several months without widespread transcriptional disruptions or cellular phenotypes. Our study thus uncovers key developmental roles for de novo H3.3 in establishing neuronal chromatin, transcriptome, identity, and connectivity immediately postmitosis that are distinct from its role in maintaining total histone H3 levels over the neuronal lifespan.
Keywords: cerebral cortex; chromatin; development; histone modification; neuronal identity.
Conflict of interest statement
The authors declare no competing interest.
Figures





References
-
- Venkatesh S., Workman J. L., Histone exchange, chromatin structure and the regulation of transcription. Nat. Rev. Mol. Cell Biol. 16, 178–189 (2015). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

