Validation of the solution structure of dimerization domain of PRC1
- PMID: 35930764
- PMCID: PMC9355583
- DOI: 10.1371/journal.pone.0270572
Validation of the solution structure of dimerization domain of PRC1
Abstract
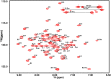
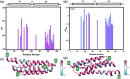
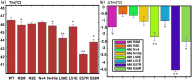
Cell-cycle dependent proteins are indispensible for the accurate division of cells, a group of proteins called Microtubule-associated proteins (MAPs) are important to cell division as it bind microtubules and participate with other co-factors to form the spindle midbody, which works as the workhorse of cell-division. PRC1 is a distinguishing member of MAPs, as it is a human MAP and works as the key in mediating daughter cell segregation in ana-phase and telo-phase. The physiological significance of PRC1 calls for a high resolution three-dimensional structure. The crystal structure of PRC1 was published but has low resolution (>3 Å) and incomplete sidechains, placing hurdles to understanding the structure-function relationships of PRC1, therefore, we determined the high-resolution solution structure of PRC1's dimerization domain using NMR spectroscopy. Significant differences between the crystal structure and the solution structure can be observed, the main differences center around the N terminus and the end of the alpha-Helix H2. Furthermore, detailed structure analyses revealed that the hydrophobic core packing of the solution and crystal structures are also different. To validate the solution structure, we used Hydrogen-deuterium exchange experiments that address the structural discrepancies between the crystal and solution structure; we also generated mutants that are key to the differences in the crystal and solution structures, measuring its structural or thermal stability by NMR spectroscopy and Fluorescence Thermal Shift Assays. These results suggest that N terminal residues are key to the integrity of the whole protein, and the solution structure of the dimerization domain better reflects the conformation PRC1 adopted in solution conditions.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures









Similar articles
-
N-Terminus-Mediated Solution Structure of Dimerization Domain of PRC1.Curr Issues Mol Biol. 2022 Apr 10;44(4):1626-1645. doi: 10.3390/cimb44040111. Curr Issues Mol Biol. 2022. PMID: 35723369 Free PMC article.
-
Marking and measuring single microtubules by PRC1 and kinesin-4.Cell. 2013 Jul 18;154(2):377-90. doi: 10.1016/j.cell.2013.06.021. Cell. 2013. PMID: 23870126 Free PMC article.
-
Insights into antiparallel microtubule crosslinking by PRC1, a conserved nonmotor microtubule binding protein.Cell. 2010 Aug 6;142(3):433-43. doi: 10.1016/j.cell.2010.07.012. Cell. 2010. PMID: 20691902 Free PMC article.
-
Mechanisms of the Ase1/PRC1/MAP65 family in central spindle assembly.Biol Rev Camb Philos Soc. 2019 Dec;94(6):2033-2048. doi: 10.1111/brv.12547. Epub 2019 Jul 25. Biol Rev Camb Philos Soc. 2019. PMID: 31343816 Review.
-
A MAP for bundling microtubules.Cell. 2010 Aug 6;142(3):364-7. doi: 10.1016/j.cell.2010.07.023. Cell. 2010. PMID: 20691897 Review.
Cited by
-
Reconciling ASPP-p53 binding mode discrepancies through an ensemble binding framework that bridges crystallography and NMR data.PLoS Comput Biol. 2024 Feb 7;20(2):e1011519. doi: 10.1371/journal.pcbi.1011519. eCollection 2024 Feb. PLoS Comput Biol. 2024. PMID: 38324587 Free PMC article.
References
-
- Jiang W, Gretchen J, Wells JN, Hope Thomas J. PRC1- A Human Mitotic Spindle?Associated CDK Substrate Protein Required for Cytokinesis.pdf. Molecular Cell. 1998;2:8. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

