Human coronaviruses: Origin, host and receptor
- PMID: 35930858
- PMCID: PMC9301904
- DOI: 10.1016/j.jcv.2022.105246
Human coronaviruses: Origin, host and receptor
Abstract
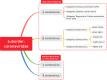
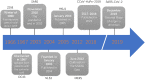
Coronavirus is a type of RNA-positive single-stranded virus with an envelope, and the spines on its surface derived its official name. Seven human coronaviruses 229E, OC43, SARS, NL63, HKU1, MERS, SARS-CoV-2 can cause both a mild cold and an epidemic of large-scale deaths and injuries. Although their clinical manifestations and many other pathogens that cause human colds are similar, studying the relationship between their evolutionary history and the receptors that infect the host can provide important insights into the natural history of human epidemics in the past and future. In this review, we describe the basic virology of these seven coronaviruses, their partial genome characteristics, and emphasize the function of receptors. We summarize the current understanding of these viruses and discuss the potential host of wild animals of these coronaviruses and the origin of zoonotic diseases.
Keywords: COVID-19; Emerging coronavirus; Host; Human coronavirus; Receptor; SARS-CoV-2.
Copyright © 2022. Published by Elsevier B.V.
Conflict of interest statement
Declaration of Competing Interest The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures

Similar articles
-
Properties of Coronavirus and SARS-CoV-2.Malays J Pathol. 2020 Apr;42(1):3-11. Malays J Pathol. 2020. PMID: 32342926 Review.
-
An overview on the seven pathogenic human coronaviruses.Rev Med Virol. 2022 Mar;32(2):e2282. doi: 10.1002/rmv.2282. Epub 2021 Aug 2. Rev Med Virol. 2022. PMID: 34339073 Review.
-
Causes and Consequences of Coronavirus Spike Protein Variability.Viruses. 2024 Jan 25;16(2):177. doi: 10.3390/v16020177. Viruses. 2024. PMID: 38399953 Free PMC article. Review.
-
Mechanistic insights into SARS-CoV-2 epidemic via revealing the features of SARS-CoV-2 coding proteins and host responses upon its infection.Bioinformatics. 2021 Jan 29;36(21):5133-5138. doi: 10.1093/bioinformatics/btaa725. Bioinformatics. 2021. PMID: 32805023 Free PMC article.
-
Spike-Independent Infection of Human Coronavirus 229E in Bat Cells.Microbiol Spectr. 2023 Jun 15;11(3):e0348322. doi: 10.1128/spectrum.03483-22. Epub 2023 May 18. Microbiol Spectr. 2023. PMID: 37199653 Free PMC article.
Cited by
-
Insights into the Activation of Unfolded Protein Response Mechanism during Coronavirus Infection.Curr Issues Mol Biol. 2024 May 5;46(5):4286-4308. doi: 10.3390/cimb46050261. Curr Issues Mol Biol. 2024. PMID: 38785529 Free PMC article. Review.
-
Vaccination against SARS-CoV-2 provides low-level cross-protection against common cold coronaviruses in mouse and non-human primate animal models.J Virol. 2025 Feb 25;99(2):e0139024. doi: 10.1128/jvi.01390-24. Epub 2025 Jan 16. J Virol. 2025. PMID: 39817773 Free PMC article.
-
Hyperforin, the major metabolite of St. John's wort, exhibits pan-coronavirus antiviral activity.Front Microbiol. 2024 Aug 8;15:1443183. doi: 10.3389/fmicb.2024.1443183. eCollection 2024. Front Microbiol. 2024. PMID: 39176276 Free PMC article.
-
COVID-19 and Diarylamidines: The Parasitic Connection.Int J Mol Sci. 2023 Apr 1;24(7):6583. doi: 10.3390/ijms24076583. Int J Mol Sci. 2023. PMID: 37047556 Free PMC article. Review.
-
Unraveling the cross-talk between a highly virulent PEDV strain and the host via single-cell transcriptomic analysis.J Virol. 2025 Jun 17;99(6):e0055525. doi: 10.1128/jvi.00555-25. Epub 2025 May 21. J Virol. 2025. PMID: 40396761 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

