DNA methyltransferase 3 alpha and TET methylcytosine dioxygenase 2 restrain mitochondrial DNA-mediated interferon signaling in macrophages
- PMID: 35931086
- PMCID: PMC9718507
- DOI: 10.1016/j.immuni.2022.06.022
DNA methyltransferase 3 alpha and TET methylcytosine dioxygenase 2 restrain mitochondrial DNA-mediated interferon signaling in macrophages
Abstract
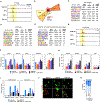
Deleterious somatic mutations in DNA methyltransferase 3 alpha (DNMT3A) and TET mehtylcytosine dioxygenase 2 (TET2) are associated with clonal expansion of hematopoietic cells and higher risk of cardiovascular disease (CVD). Here, we investigated roles of DNMT3A and TET2 in normal human monocyte-derived macrophages (MDM), in MDM isolated from individuals with DNMT3A or TET2 mutations, and in macrophages isolated from human atherosclerotic plaques. We found that loss of function of DNMT3A or TET2 resulted in a type I interferon response due to impaired mitochondrial DNA integrity and activation of cGAS signaling. DNMT3A and TET2 normally maintained mitochondrial DNA integrity by regulating the expression of transcription factor A mitochondria (TFAM) dependent on their interactions with RBPJ and ZNF143 at regulatory regions of the TFAM gene. These findings suggest that targeting the cGAS-type I IFN pathway may have therapeutic value in reducing risk of CVD in patients with DNMT3A or TET2 mutations.
Keywords: DNMT3A; TET2; TFAM; atherosclerosis; clonal hematopoiesis; interferon; mitochondria DNA; transcriptional regulation.
Copyright © 2022 Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests C.K.G. is a cofounder and member of the scientific advisory board of Asteroid Therapeutics.
Figures


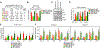



Comment in
-
Clonal hematopoiesis, inflammation, and cardiovascular disorders: a mitochondrial connection.Trends Immunol. 2022 Sep;43(9):693-695. doi: 10.1016/j.it.2022.07.009. Epub 2022 Aug 6. Trends Immunol. 2022. PMID: 35945112
References
-
- Acuna-Hidalgo R, Sengul H, Steehouwer M, van de Vorst M, Vermeulen SH, Kiemeney L, Veltman JA, Gilissen C, and Hoischen A (2017). Ultra-sensitive sequencing identifies high prevalence of clonal hematopoiesis-associated mutations throughout adult life. Am. J. Hum. Genet 101, 50–64. 10.1016/j.ajhg.2017.05.013. - DOI - PMC - PubMed
-
- Assmus B, Cremer S, Kirschbaum K, Culmann D, Kiefer K, Dorsheimer L, Rasper T, Abou-El-Ardat K, Herrmann E, Berkowitsch A, et al. (2021). Clonal haematopoiesis in chronic ischaemic heart failure: prognostic role of clone size for DNMT3A- and TET2-driver gene mutations. Eur. Heart J 42, 257–265. 10.1093/eurheartj/ehaa845. - DOI - PubMed
-
- Buscarlet M, Provost S, Zada YF, Barhdadi A, Bourgoin V, Lépine G, Mollica L, Szuber N, Dubé MP, and Busque L (2017). DNMT3A and TET2 dominate clonal hematopoiesis and demonstrate benign phenotypes and different genetic predispositions. Blood 130, 753–762. 10.1182/blood-2017-04-777029. - DOI - PubMed

