Pre-existing partner-drug resistance to artemisinin combination therapies facilitates the emergence and spread of artemisinin resistance: a consensus modelling study
- PMID: 35931099
- PMCID: PMC9436785
- DOI: 10.1016/S2666-5247(22)00155-0
Pre-existing partner-drug resistance to artemisinin combination therapies facilitates the emergence and spread of artemisinin resistance: a consensus modelling study
Erratum in
-
Correction to Lancet Microbe 2022; 3: e701-10.Lancet Microbe. 2023 Jan;4(1):e17. doi: 10.1016/S2666-5247(22)00353-6. Epub 2022 Nov 25. Lancet Microbe. 2023. PMID: 36442493 Free PMC article. No abstract available.
Abstract
Background: Artemisinin-resistant genotypes of Plasmodium falciparum have now emerged a minimum of six times on three continents despite recommendations that all artemisinins be deployed as artemisinin combination therapies (ACTs). Widespread resistance to the non-artemisinin partner drugs in ACTs has the potential to limit the clinical and resistance benefits provided by combination therapy. We aimed to model and evaluate the long-term effects of high levels of partner-drug resistance on the early emergence of artemisinin-resistant genotypes.
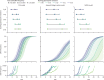
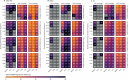
Methods: Using a consensus modelling approach, we used three individual-based mathematical models of Plasmodium falciparum transmission to evaluate the effects of pre-existing partner-drug resistance and ACT deployment on the evolution of artemisinin resistance. Each model simulates 100 000 individuals in a particular transmission setting (malaria prevalence of 1%, 5%, 10%, or 20%) with a daily time step that updates individuals' infection status, treatment status, immunity, genotype-specific parasite densities, and clinical state. We modelled varying access to antimalarial drugs if febrile (coverage of 20%, 40%, or 60%) with one primary ACT used as first-line therapy: dihydroartemisinin-piperaquine (DHA-PPQ), artesunate-amodiaquine (ASAQ), or artemether-lumefantrine (AL). The primary outcome was time until 0·25 580Y allele frequency for artemisinin resistance (the establishment time).
Findings: Higher frequencies of pre-existing partner-drug resistant genotypes lead to earlier establishment of artemisinin resistance. Across all models, a 10-fold increase in the frequency of partner-drug resistance genotypes on average corresponded to loss of artemisinin efficacy 2-12 years earlier. Most reductions in time to artemisinin resistance establishment were observed after an increase in frequency of the partner-drug resistance genotype from 0·0 to 0·10.
Interpretation: Partner-drug resistance in ACTs facilitates the early emergence of artemisinin resistance and is a major public health concern. Higher-grade partner-drug resistance has the largest effect, with piperaquine resistance accelerating the early emergence of artemisinin-resistant alleles the most. Continued investment in molecular surveillance of partner-drug resistant genotypes to guide choice of first-line ACT is paramount.
Funding: Schmidt Science Fellowship in partnership with the Rhodes Trust; Bill & Melinda Gates Foundation; Wellcome Trust.
Copyright © 2022 The Author(s). Published by Elsevier Ltd. This is an Open Access article under the CC BY 4.0 license. Published by Elsevier Ltd.. All rights reserved.
Conflict of interest statement
Declaration of interests We declare no competing interests.
Figures


References
-
- WHO . 1st edition. World Health Organization; Geneva: 2006. Guidelines for the treatment of malaria.
-
- WHO WHO briefing on malaria treatment guidelines and artemisinin monotherapies. April 19, 2006. http://www.malaria.org/ABOUT%20MALARIA/Meeting_briefing19April%20on%20Tr...
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

