Extensive genome introgression between domestic ferret and European polecat during population recovery in Great Britain
- PMID: 35932226
- PMCID: PMC9584812
- DOI: 10.1093/jhered/esac038
Extensive genome introgression between domestic ferret and European polecat during population recovery in Great Britain
Abstract
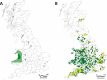
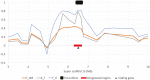
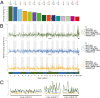
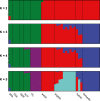
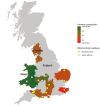
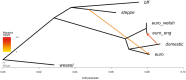
The European polecat (Mustela putorius) is a mammalian predator which occurs across much of Europe east to the Ural Mountains. In Great Britain, following years of persecution the range of the European polecat contracted and by the early 1900s was restricted to unmanaged forests of central Wales. The European polecat has recently undergone a population increase due to legal protection and its range now overlaps that of feral domestic ferrets (Mustela putorius furo). During this range expansion, European polecats hybridized with feral domestic ferrets producing viable offspring. Here, we carry out population-level whole-genome sequencing on 8 domestic ferrets, 19 British European polecats, and 15 European polecats from the European mainland. We used a range of population genomics methods to examine the data, including phylogenetics, phylogenetic graphs, model-based clustering, phylogenetic invariants, ABBA-BABA tests, topology weighting, and Fst. We found high degrees of genome introgression in British polecats outside their previous stronghold, even in those individuals phenotyped as "pure" polecats. These polecats ranged from presumed F1 hybrids (gamma = 0.53) to individuals that were much less introgressed (gamma = 0.2). We quantify this introgression and find introgressed genes containing Fst outliers associated with cognitive function and sight.
Keywords: European polecat; Mustelids; conservation; domestic ferret; genomics; introgression.
© The American Genetic Association. 2022.
Conflict of interest statement
None declared.
Figures


References
-
- Aleo S, et al. Autism spectrum disorder and intellectual disability in an inherited 2q14.3 micro-deletion involving CNTNAP5. Am J Med Genet A. 2020;182(12):3071–3073. - PubMed
-
- Alves PC, et al. Evidence for genetic similarity of two allopatric European hares (Lepus corsicanus and L. castroviejoi) inferred from nuclear DNA sequences. Mol Phylogenet Evol. 2008;46(3):1191–1197. - PubMed
-
- Andrews, S. 2010. FastQC: a quality control tool for high throughput sequence data. Available online at: http://www.bioinformatics.babraham.ac.uk/projects/fastqc/
-
- Birks J. Polecats. Whittet Books,Stansted, United Kingdom; 2015.
-
- Birks J, Kitchener A. The distribution and status of the polecat Mustela putorius in Britain in the 1990s. Vol. 152. London: The Vincent Wildlife Trust; 1999.
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

