Characterization of two kdr mutations at predicted pyrethroid receptor site 2 in the sodium channels of Aedes aegypti and Nilaparvata lugens
- PMID: 35932971
- PMCID: PMC10076083
- DOI: 10.1016/j.ibmb.2022.103814
Characterization of two kdr mutations at predicted pyrethroid receptor site 2 in the sodium channels of Aedes aegypti and Nilaparvata lugens
Abstract
Pyrethroid insecticides prolong the opening of insect sodium channels by binding to two predicted pyrethroid receptor sites (PyR), PyR1 and PyR2. Many naturally-occurring sodium channel mutations that confer pyrethroid resistance (known as knockdown resistance, kdr) are located at PyR1. Recent studies identified two new mutations, V253F and T267A, at PyR2, which co-exist with two well-known mutations F1534C or M918T, at PyR1, in pyrethroid-resistant populations of Aedes aegypti and Nilaparvata lugens, respectively. However, the role of the V253F and T267A mutations in pyrethroid resistance has not been functionally examined. Here we report functional characterization of the V253F and T267A mutations in the Ae. aegypti sodium channel AaNav2-1 and the N. lugens sodium channel NlNav1 expressed in Xenopus oocytes. Both mutations alone reduced channel sensitivity to pyrethroids, including etofenprox. We docked etofenprox in a homology model of the pore module of the NlNav1 channel based on the crystal structure of an open prokaryotic sodium channel NavMs. In the low-energy binding pose etofenprox formed contacts with V253, T267 and a previously identified L1014 within PyR2. Combining of V253F or T267A with F1534C or M918T results in a higher level of pyrethroid insensitivity. Furthermore, both V253F and T267A mutations altered channel gating properties. However, V253F- and T267A-induced gating modifications was not observed in the double mutant channels. Our findings highlight the first example in which naturally-found combinational mutations in PyR1 and PyR2 not only confer higher level pyrethroid insensitivity, but also reduce potential fitness tradeoff in pyrethroid-resistant mosquitoes caused by kdr mutation-induced sodium channel gating modifications.
Keywords: Knockdown resistance; Pyrethroid insecticides; Pyrethroid receptor site 2; Sodium channel.
Copyright © 2022 Elsevier Ltd. All rights reserved.
Figures

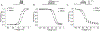

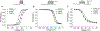
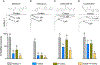

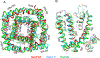

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

