A conserved sequence motif in the Escherichia coli soluble FAD-containing pyridine nucleotide transhydrogenase is important for reaction efficiency
- PMID: 35933012
- PMCID: PMC9460512
- DOI: 10.1016/j.jbc.2022.102304
A conserved sequence motif in the Escherichia coli soluble FAD-containing pyridine nucleotide transhydrogenase is important for reaction efficiency
Abstract
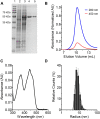
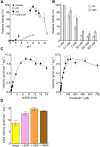
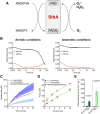
Soluble pyridine nucleotide transhydrogenases (STHs) are flavoenzymes involved in the redox homeostasis of the essential cofactors NAD(H) and NADP(H). They catalyze the reversible transfer of reducing equivalents between the two nicotinamide cofactors. The soluble transhydrogenase from Escherichia coli (SthA) has found wide use in both in vivo and in vitro applications to steer reducing equivalents toward NADPH-requiring reactions. However, mechanistic insight into SthA function is still lacking. In this work, we present a biochemical characterization of SthA, focusing for the first time on the reactivity of the flavoenzyme with molecular oxygen. We report on oxidase activity of SthA that takes place both during transhydrogenation and in the absence of an oxidized nicotinamide cofactor as an electron acceptor. We find that this reaction produces the reactive oxygen species hydrogen peroxide and superoxide anion. Furthermore, we explore the evolutionary significance of the well-conserved CXXXXT motif that distinguishes STHs from the related family of flavoprotein disulfide reductases in which a CXXXXC motif is conserved. Our mutational analysis revealed the cysteine and threonine combination in SthA leads to better coupling efficiency of transhydrogenation and reduced reactive oxygen species release compared to enzyme variants with mutated motifs. These results expand our mechanistic understanding of SthA by highlighting reactivity with molecular oxygen and the importance of the evolutionarily conserved sequence motif.
Keywords: flavoprotein; nicotinamide cofactors; protein engineering; reactive oxygen species; soluble transhydrogenase.
Copyright © 2022 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Conflict of interest The authors declare that there are no conflicts of interest with the contents of this article.
Figures

Similar articles
-
A spontaneous mutation in the nicotinamide nucleotide transhydrogenase gene of C57BL/6J mice results in mitochondrial redox abnormalities.Free Radic Biol Med. 2013 Oct;63:446-56. doi: 10.1016/j.freeradbiomed.2013.05.049. Epub 2013 Jun 7. Free Radic Biol Med. 2013. PMID: 23747984
-
The mechanism of hydride transfer between NADH and 3-acetylpyridine adenine dinucleotide by the pyridine nucleotide transhydrogenase of Escherichia coli.Biochim Biophys Acta. 1995 Oct 10;1231(3):297-303. doi: 10.1016/0005-2728(95)00089-2. Biochim Biophys Acta. 1995. PMID: 7578217
-
Cofactor regeneration by a soluble pyridine nucleotide transhydrogenase for biological production of hydromorphone.Appl Environ Microbiol. 2000 Dec;66(12):5161-6. doi: 10.1128/AEM.66.12.5161-5166.2000. Appl Environ Microbiol. 2000. PMID: 11097884 Free PMC article.
-
Domains, specific residues and conformational states involved in hydride ion transfer and proton pumping by nicotinamide nucleotide transhydrogenase from Escherichia coli.Biochim Biophys Acta. 1998 Jun 10;1365(1-2):10-6. doi: 10.1016/s0005-2728(98)00038-3. Biochim Biophys Acta. 1998. PMID: 9693716 Review.
-
Nicotinamide nucleotide transhydrogenase: a model for utilization of substrate binding energy for proton translocation.FASEB J. 1996 Mar;10(4):444-52. doi: 10.1096/fasebj.10.4.8647343. FASEB J. 1996. PMID: 8647343 Review.
Cited by
-
CryoEM-enabled visual proteomics reveals de novo structures of oligomeric protein complexes.Structure. 2025 Jul 8:S0969-2126(25)00223-0. doi: 10.1016/j.str.2025.06.007. Online ahead of print. Structure. 2025. PMID: 40664216
-
A Hitchhiker's Guide to Supplying Enzymatic Reducing Power into Synthetic Cells.ACS Synth Biol. 2023 Apr 21;12(4):947-962. doi: 10.1021/acssynbio.3c00070. Epub 2023 Apr 13. ACS Synth Biol. 2023. PMID: 37052416 Free PMC article. Review.
-
Toehold Switch-Based Approach for Engineering Acid-Tolerance Modules to Enhance Production Robustness of Industrial E. coli Strains at Low pH.Microb Biotechnol. 2025 Jun;18(6):e70175. doi: 10.1111/1751-7915.70175. Microb Biotechnol. 2025. PMID: 40485106 Free PMC article.
References
-
- Blank L.M., Ebert B.E., Buehler K., Bühler B. Redox biocatalysis and metabolism: molecular mechanisms and metabolic network analysis. Antioxid. Redox Signal. 2010;13:349–394. - PubMed
-
- Jackson J.B. Proton translocation by transhydrogenase. FEBS Lett. 2003;545:18–24. - PubMed
-
- Argyrou A., Blanchard J.S. Flavoprotein disulfide reductases: advances in chemistry and function. Prog. Nucl. Acid Res. Mol. Biol. 2004;78:89–142. - PubMed
-
- Kleiger G., Eisenberg D. GXXXG and GXXXA motifs stabilize FAD and NAD(P)-binding rossmann folds through Cα-H···O hydrogen bonds and van der waals interactions. J. Mol. Biol. 2002;323:69–76. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

