Oxygen-dependent regulation of E3(SCF)ubiquitin ligases and a Skp1-associated JmjD6 homolog in development of the social amoeba Dictyostelium
- PMID: 35933019
- PMCID: PMC9485057
- DOI: 10.1016/j.jbc.2022.102305
Oxygen-dependent regulation of E3(SCF)ubiquitin ligases and a Skp1-associated JmjD6 homolog in development of the social amoeba Dictyostelium
Abstract
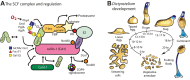
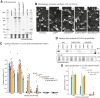
E3-SCF (Skp1/cullin-1/F-box protein) polyubiquitin ligases activate the proteasomal degradation of over a thousand proteins, but the evolutionary diversification of the F-box protein (FBP) family of substrate receptor subunits has challenged their elucidation in protists. Here, we expand the FBP candidate list in the social amoeba Dictyostelium and show that the Skp1 interactome is highly remodeled as cells transition from growth to multicellular development. Importantly, a subset of candidate FBPs was less represented when the posttranslational hydroxylation and glycosylation of Skp1 was abrogated by deletion of the O2-sensing Skp1 prolyl hydroxylase PhyA. A role for this Skp1 modification for SCF activity was indicated by partial rescue of development, which normally depends on high O2 and PhyA, of phyA-KO cells by proteasomal inhibitors. Further examination of two FBPs, FbxwD and the Jumonji C protein JcdI, suggested that Skp1 was substituted by other factors in phyA-KO cells. Although a double-KO of jcdI and its paralog jcdH did not affect development, overexpression of JcdI increased its sensitivity to O2. JcdI, a nonheme dioxygenase shown to have physiological O2 dependence, is conserved across protists with its F-box and other domains, and is related to the human oncogene JmjD6. Sensitization of JcdI-overexpression cells to O2 depended on its dioxygenase activity and other domains, but not its F-box, which may however be the mediator of its reduced levels in WT relative to Skp1 modification mutant cells. The findings suggest that activation of JcdI by O2 is tempered by homeostatic downregulation via PhyA and association with Skp1.
Keywords: E3(SCF)ubiquitin-ligase; F-box protein; Jumonji C; Skp1; cellular slime mold; glycosylation; oxygen; prolyl hydroxylase.
Copyright © 2022 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Conflict of interest The authors declare that they have no conflicts of interest with the contents of this article.
Figures






References
-
- Willems A.R., Schwab M., Tyers M. A hitchhiker's guide to the cullin ubiquitin ligases: SCF and its kin. Biochim. Biophys. Acta - Mol. Cell Res. 2004;1695:133–170. - PubMed
-
- Schulman B.A., Carrano A.C., Jeffrey P.D., Bowen Z., Kinnucan E.R., Finnin M.S., et al. Insights into SCF ubiquitin ligases from the structure of the Skp1-Skp2 complex. Nature. 2000;408:381–386. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

