Improved prediction of gene expression through integrating cell signalling models with machine learning
- PMID: 35933367
- PMCID: PMC9356471
- DOI: 10.1186/s12859-022-04787-8
Improved prediction of gene expression through integrating cell signalling models with machine learning
Abstract
Background: A key problem in bioinformatics is that of predicting gene expression levels. There are two broad approaches: use of mechanistic models that aim to directly simulate the underlying biology, and use of machine learning (ML) to empirically predict expression levels from descriptors of the experiments. There are advantages and disadvantages to both approaches: mechanistic models more directly reflect the underlying biological causation, but do not directly utilize the available empirical data; while ML methods do not fully utilize existing biological knowledge.
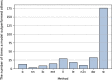
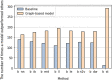
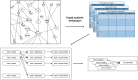
Results: Here, we investigate overcoming these disadvantages by integrating mechanistic cell signalling models with ML. Our approach to integration is to augment ML with similarity features (attributes) computed from cell signalling models. Seven sets of different similarity feature were generated using graph theory. Each set of features was in turn used to learn multi-target regression models. All the features have significantly improved accuracy over the baseline model - without the similarity features. Finally, the seven multi-target regression models were stacked together to form an overall prediction model that was significantly better than the baseline on 95% of genes on an independent test set. The similarity features enable this stacking model to provide interpretable knowledge about cancer, e.g. the role of ERBB3 in the MCF7 breast cancer cell line.
Conclusion: Integrating mechanistic models as graphs helps to both improve the predictive results of machine learning models, and to provide biological knowledge about genes that can help in building state-of-the-art mechanistic models.
Keywords: Gene expression; Machine learning; Multi-target regression.
© 2022. The Author(s).
Conflict of interest statement
We have no competing interests.
Figures

References
-
- Richardson LF. Weather prediction by numerical process. Cambridge: Cambridge University Press; 2007.
-
- Grover A, Kapoor A, Horvitz E. A deep hybrid model for weather forecasting. In: Proceedings of the 21th ACM SIGKDD international conference on knowledge discovery and data mining; 2015. pp. 379–386.
-
- Kryshtafovych A, Schwede T, Topf M, Fidelis K, Moult J. Critical assessment of methods of protein structure prediction (CASP)—Round XIV. Proteins: Structure, Function, and Bioinformatics; n/a(n/a). https://onlinelibrary.wiley.com/doi/abs/10.1002/prot.26237. - DOI - PMC - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials

