USP22 controls type III interferon signaling and SARS-CoV-2 infection through activation of STING
- PMID: 35933402
- PMCID: PMC9357023
- DOI: 10.1038/s41419-022-05124-w
USP22 controls type III interferon signaling and SARS-CoV-2 infection through activation of STING
Abstract
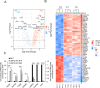
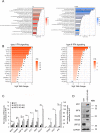
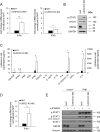
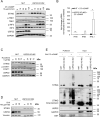
Pattern recognition receptors (PRRs) and interferons (IFNs) serve as essential antiviral defense against SARS-CoV-2, the causative agent of the COVID-19 pandemic. Type III IFNs (IFN-λ) exhibit cell-type specific and long-lasting functions in auto-inflammation, tumorigenesis, and antiviral defense. Here, we identify the deubiquitinating enzyme USP22 as central regulator of basal IFN-λ secretion and SARS-CoV-2 infections in human intestinal epithelial cells (hIECs). USP22-deficient hIECs strongly upregulate genes involved in IFN signaling and viral defense, including numerous IFN-stimulated genes (ISGs), with increased secretion of IFN-λ and enhanced STAT1 signaling, even in the absence of exogenous IFNs or viral infection. Interestingly, USP22 controls basal and 2'3'-cGAMP-induced STING activation and loss of STING reversed STAT activation and ISG and IFN-λ expression. Intriguingly, USP22-deficient hIECs are protected against SARS-CoV-2 infection, viral replication, and the formation of de novo infectious particles, in a STING-dependent manner. These findings reveal USP22 as central host regulator of STING and type III IFN signaling, with important implications for SARS-CoV-2 infection and antiviral defense.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures


Similar articles
-
Antiviral Activity of Type I, II, and III Interferons Counterbalances ACE2 Inducibility and Restricts SARS-CoV-2.mBio. 2020 Sep 10;11(5):e01928-20. doi: 10.1128/mBio.01928-20. mBio. 2020. PMID: 32913009 Free PMC article.
-
Increased Sensitivity of SARS-CoV-2 to Type III Interferon in Human Intestinal Epithelial Cells.J Virol. 2022 Apr 13;96(7):e0170521. doi: 10.1128/jvi.01705-21. Epub 2022 Mar 9. J Virol. 2022. PMID: 35262371 Free PMC article.
-
Inhibition of SARS-CoV-2 by type I and type III interferons.J Biol Chem. 2020 Oct 9;295(41):13958-13964. doi: 10.1074/jbc.AC120.013788. Epub 2020 Jun 25. J Biol Chem. 2020. PMID: 32587093 Free PMC article.
-
Interferon-lambda: New role in intestinal symptoms of COVID-19.World J Gastroenterol. 2023 Apr 7;29(13):1942-1954. doi: 10.3748/wjg.v29.i13.1942. World J Gastroenterol. 2023. PMID: 37155525 Free PMC article. Review.
-
Type I and III interferon responses in SARS-CoV-2 infection.Exp Mol Med. 2021 May;53(5):750-760. doi: 10.1038/s12276-021-00592-0. Epub 2021 May 6. Exp Mol Med. 2021. PMID: 33953323 Free PMC article. Review.
Cited by
-
Hycanthone Inhibits Inflammasome Activation and Neuroinflammation-Induced Depression-Like Behaviors in Mice.Biomol Ther (Seoul). 2023 Mar 1;31(2):161-167. doi: 10.4062/biomolther.2022.073. Epub 2022 Oct 7. Biomol Ther (Seoul). 2023. PMID: 36203404 Free PMC article.
-
Usp22 Deficiency Leads to Downregulation of PD-L1 and Pathological Activation of CD8+ T Cells and Causes Immunopathology in Response to Acute LCMV Infection.Vaccines (Basel). 2023 Oct 5;11(10):1563. doi: 10.3390/vaccines11101563. Vaccines (Basel). 2023. PMID: 37896966 Free PMC article.
-
In search of a function for human type III interferons: insights from inherited and acquired deficits.Curr Opin Immunol. 2024 Apr;87:102427. doi: 10.1016/j.coi.2024.102427. Epub 2024 May 22. Curr Opin Immunol. 2024. PMID: 38781720 Free PMC article. Review.
-
The Dual Role of cGAS-STING Signaling in COVID-19: Implications for Therapy.Cells. 2025 Feb 28;14(5):362. doi: 10.3390/cells14050362. Cells. 2025. PMID: 40072090 Free PMC article. Review.
-
Progress of cGAS-STING signaling in response to SARS-CoV-2 infection.Front Immunol. 2022 Dec 7;13:1010911. doi: 10.3389/fimmu.2022.1010911. eCollection 2022. Front Immunol. 2022. PMID: 36569852 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

