shRNA‑mediated knockdown of KNTC1 inhibits non-small-cell lung cancer through regulating PSMB8
- PMID: 35933405
- PMCID: PMC9357013
- DOI: 10.1038/s41419-022-05140-w
shRNA‑mediated knockdown of KNTC1 inhibits non-small-cell lung cancer through regulating PSMB8
Abstract
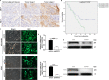
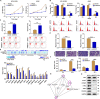
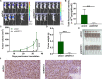
In view of the important roles played by Kinetochore proteins in mitosis, we believed that they may contribute to the development and progression of human cancers, which has been reported recently elsewhere. Kinetochore-associated 1 (KNTC1) participates in the segregation of sister chromatids during mitosis, the effects of which on non-small-cell lung cancer (NSCLC) remain unclear. Here, we sought to identify the biological significance of KNTC1 in NSCLC. KNTC1 protein expression in NSCLC tissues was investigated by immunohistochemistry. Lentivirus delivered short hairpin RNA (shRNA) was utilized to establish KNTC1 silence NSCLC cell lines. The effects of KNTC1 depletion on NSCLC cell proliferation, migration, apoptosis, and tumor formation were analyzed by MTT assay, wound-healing assay, transwell assay, flow cytometry assay, and in nude mouse models in vivo. After KNTC1 reduction, NSCLC cell viability, proliferation, migration, and invasion were restrained. A xenograft tumor model was also provided to demonstrate the inhibited tumorigenesis in NSCLC. In addition, the downstream mechanism analysis indicated that KNTC1 depletion was positively associated with PSMB8. The findings of the present study suggested that KNTC1 may have a pivotal role in mediating NSCLC progression and may act as a novel therapeutic target for NSCLC.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures


Similar articles
-
shRNA‑mediated knockdown of KNTC1 suppresses cell viability and induces apoptosis in esophageal squamous cell carcinoma.Int J Oncol. 2019 Mar;54(3):1053-1060. doi: 10.3892/ijo.2019.4672. Epub 2019 Jan 3. Int J Oncol. 2019. PMID: 30628654
-
Silencing of KNTC1 inhibits hepatocellular carcinoma cells progression via suppressing PI3K/Akt pathway.Cell Signal. 2023 Jan;101:110498. doi: 10.1016/j.cellsig.2022.110498. Epub 2022 Oct 21. Cell Signal. 2023. PMID: 36273753
-
Long noncoding RNA PSMA3‑AS1 functions as a microRNA‑409‑3p sponge to promote the progression of non‑small cell lung carcinoma by targeting spindlin 1.Oncol Rep. 2020 Oct;44(4):1550-1560. doi: 10.3892/or.2020.7693. Epub 2020 Jul 15. Oncol Rep. 2020. PMID: 32945481 Free PMC article.
-
Knockdown of LncRNA PVT1 inhibits tumorigenesis in non-small-cell lung cancer by regulating miR-497 expression.Exp Cell Res. 2018 Jan 1;362(1):172-179. doi: 10.1016/j.yexcr.2017.11.014. Epub 2017 Nov 11. Exp Cell Res. 2018. PMID: 29133127
-
Kinetochore-associated protein 1 promotes the invasion and tumorigenicity of cervical cancer cells via matrix metalloproteinase-2 and matrix metalloproteinase-9.Bioengineered. 2022 Apr;13(4):9495-9507. doi: 10.1080/21655979.2022.2061144. Bioengineered. 2022. PMID: 35389773 Free PMC article.
Cited by
-
Applications and advancements of nanoparticle-based drug delivery in alleviating lung cancer and chronic obstructive pulmonary disease.Naunyn Schmiedebergs Arch Pharmacol. 2024 May;397(5):2793-2833. doi: 10.1007/s00210-023-02830-w. Epub 2023 Nov 22. Naunyn Schmiedebergs Arch Pharmacol. 2024. PMID: 37991539 Review.
-
FAM50A as a novel prognostic marker modulates the proliferation of colorectal cancer cells via CylinA2/CDK2 pathway.PLoS One. 2025 Feb 25;20(2):e0318776. doi: 10.1371/journal.pone.0318776. eCollection 2025. PLoS One. 2025. PMID: 39999107 Free PMC article.
-
KNTC1 knockdown inhibits proliferation and metastases of liver cancer.3 Biotech. 2023 Sep;13(9):309. doi: 10.1007/s13205-023-03722-9. Epub 2023 Aug 22. 3 Biotech. 2023. PMID: 37621322 Free PMC article.
-
Potential 'anti-cancer' effects of esketamine on proliferation, apoptosis, migration and invasion in esophageal squamous carcinoma cells.Eur J Med Res. 2023 Nov 15;28(1):517. doi: 10.1186/s40001-023-01511-x. Eur J Med Res. 2023. PMID: 37968758 Free PMC article.
-
Multifaceted investigations of PSMB8 provides insights into prognostic prediction and immunological target in thyroid carcinoma.PLoS One. 2025 May 7;20(5):e0323013. doi: 10.1371/journal.pone.0323013. eCollection 2025. PLoS One. 2025. PMID: 40334200 Free PMC article.
References
-
- Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424. - PubMed
-
- Miller KD, Nogueira L, Mariotto AB, Rowland JH, Siegel RL Cancer treatment and survivorship statistics, 2019. CA Cancer J Clin. 2019;69:363–85. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

