Temporal profiling of the breast tumour microenvironment reveals collagen XII as a driver of metastasis
- PMID: 35933466
- PMCID: PMC9357007
- DOI: 10.1038/s41467-022-32255-7
Temporal profiling of the breast tumour microenvironment reveals collagen XII as a driver of metastasis
Abstract
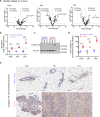
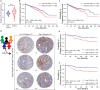
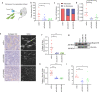
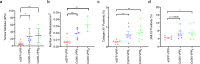
The tumour stroma, and in particular the extracellular matrix (ECM), is a salient feature of solid tumours that plays a crucial role in shaping their progression. Many desmoplastic tumours including breast cancer involve the significant accumulation of type I collagen. However, recently it has become clear that the precise distribution and organisation of matrix molecules such as collagen I is equally as important in the tumour as their abundance. Cancer-associated fibroblasts (CAFs) coexist within breast cancer tissues and play both pro- and anti-tumourigenic roles through remodelling the ECM. Here, using temporal proteomic profiling of decellularized tumours, we interrogate the evolving matrisome during breast cancer progression. We identify 4 key matrisomal clusters, and pinpoint collagen type XII as a critical component that regulates collagen type I organisation. Through combining our proteomics with single-cell transcriptomics, and genetic manipulation models, we show how CAF-secreted collagen XII alters collagen I organisation to create a pro-invasive microenvironment supporting metastatic dissemination. Finally, we show in patient cohorts that collagen XII may represent an indicator of breast cancer patients at high risk of metastatic relapse.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures





References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

