Leptin mutation and mycobacterial infection lead non-synergistically to a similar metabolic syndrome
- PMID: 35933481
- PMCID: PMC9356939
- DOI: 10.1007/s11306-022-01921-8
Leptin mutation and mycobacterial infection lead non-synergistically to a similar metabolic syndrome
Abstract
Introduction: The leptin signaling pathway plays an important role as a key regulator of glucose homeostasis, metabolism control and systemic inflammatory responses. However, the metabolic effects of leptin on infectious diseases, for example tuberculosis (TB), are still little known.
Objectives: In this study, we aim to investigate the role of leptin on metabolism in the absence and presence of mycobacterial infection in zebrafish larvae and mice.
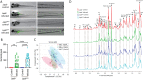
Methods: Metabolites in entire zebrafish larvae and the blood of mice were studied using high-resolution magic-angle-spinning nuclear magnetic resonance (HR-MAS NMR) spectroscopy and mass spectrometry, respectively. For transcriptome studies of zebrafish larvae, deep RNA sequencing was used.
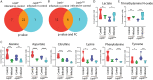
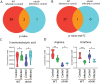
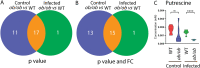
Results: The results show that leptin mutation leads to a similar metabolic syndrome as caused by mycobacterial infection in the two species, characterized by the decrease of 11 amine metabolites. In both species, this metabolic syndrome was not aggravated further when the leptin mutant was infected by mycobacteria. Therefore, we conclude that leptin and mycobacterial infection are both impacting metabolism non-synergistically. In addition, we studied the transcriptomes of lepbibl54 mutant zebrafish larvae and wild type (WT) siblings after mycobacterial infection. These studies showed that mycobacteria induced a very distinct transcriptome signature in the lepbibl54 mutant zebrafish compared to WT sibling control larvae. Furthermore, lepbibl55 Tg (pck1:luc1) zebrafish line was constructed and confirmed this difference in transcriptional responses.
Conclusions: Leptin mutation and TB lead non-synergistically to a similar metabolic syndrome. Moreover, different transcriptomic responses in the lepbibl54 mutant and TB can lead to the similar metabolic end states.
Keywords: Leptin mutant zebrafish; Metabolomics; Non-synergy; Ob/ob mice; Transcriptomics; Tuberculosis.
© 2022. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests.
Figures



References
-
- Andrade BB, Kumar NP, Sridhar R, Banurekha VV, Jawahar MS, Nutman TB, Sher A, Babu S. Heightened plasma levels of heme oxygenase-1 and tissue inhibitor of metalloproteinase-4 as well as elevated peripheral neutrophil counts are associated with TB-diabetes comorbidity. Chest. 2014;145:1244–1254. doi: 10.1378/chest.13-1799. - DOI - PMC - PubMed
-
- Deng J, Liu L, Yang Q, Wei C, Zhang H, Xin H, Pan S, Liu Z, Wang D, Liu B, Gao L, Liu R, Pang Y, Chen X, Zheng J, Jin Q. Urinary metabolomic analysis to identify potential markers for the diagnosis of tuberculosis and latent tuberculosis. Archives of Biochemistry and Biophysics. 2021;704:108876. doi: 10.1016/j.abb.2021.108876. - DOI - PubMed
-
- Ding Y, Haks MC, Forn-Cuní G, He J, Nowik N, Harms AC, Hankemeier T, Eeza MNH, Matysik J, Alia A, Spaink HP. Metabolomic and transcriptomic profiling of adult mice and larval zebrafish leptin mutants reveal a common pattern of changes in metabolites and signaling pathways. Cell and Bioscience. 2021;11:126. doi: 10.1186/s13578-021-00642-0. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous
